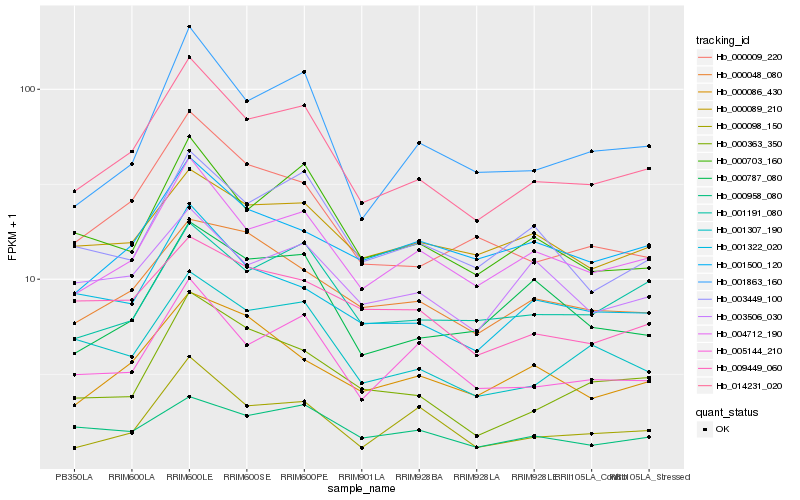
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_014231_020 |
0.0 |
- |
- |
PREDICTED: ras-related protein Rab11D [Jatropha curcas] |
| 2 |
Hb_004712_190 |
0.070565573 |
- |
- |
PREDICTED: uncharacterized protein LOC105633105 [Jatropha curcas] |
| 3 |
Hb_001863_160 |
0.0893824029 |
- |
- |
PREDICTED: 14-3-3-like protein [Jatropha curcas] |
| 4 |
Hb_000703_160 |
0.0921324319 |
- |
- |
PREDICTED: serine/threonine-protein kinase EDR1 isoform X2 [Jatropha curcas] |
| 5 |
Hb_005144_210 |
0.0922312364 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_000009_220 |
0.0933491199 |
- |
- |
PREDICTED: calcium uniporter protein 2, mitochondrial-like [Jatropha curcas] |
| 7 |
Hb_001307_190 |
0.0966458796 |
transcription factor |
TF Family: G2-like |
transcription factor, putative [Ricinus communis] |
| 8 |
Hb_001191_080 |
0.0973969317 |
- |
- |
protein with unknown function [Ricinus communis] |
| 9 |
Hb_001322_020 |
0.1010728938 |
- |
- |
PREDICTED: polycomb group protein VERNALIZATION 2-like isoform X2 [Populus euphratica] |
| 10 |
Hb_003449_100 |
0.1038924674 |
- |
- |
PREDICTED: protein YIPF6 homolog [Jatropha curcas] |
| 11 |
Hb_000048_080 |
0.1062012294 |
- |
- |
PREDICTED: cyclin-dependent kinase D-3 isoform X1 [Jatropha curcas] |
| 12 |
Hb_000363_350 |
0.107639975 |
- |
- |
PREDICTED: transcription initiation factor TFIID subunit 6-like [Fragaria vesca subsp. vesca] |
| 13 |
Hb_000958_080 |
0.1098982925 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_000787_080 |
0.1111466055 |
- |
- |
PREDICTED: TBC1 domain family member 16-like [Jatropha curcas] |
| 15 |
Hb_000086_430 |
0.1120121312 |
- |
- |
hypothetical protein JCGZ_17649 [Jatropha curcas] |
| 16 |
Hb_003506_030 |
0.1122920634 |
- |
- |
PREDICTED: ATP-dependent Clp protease ATP-binding subunit clpX-like, mitochondrial isoform X2 [Jatropha curcas] |
| 17 |
Hb_009449_060 |
0.1129605127 |
- |
- |
PREDICTED: ubiquitin-like-specific protease 1D isoform X2 [Jatropha curcas] |
| 18 |
Hb_000098_150 |
0.1145232749 |
- |
- |
DNA helicase hus2, putative [Ricinus communis] |
| 19 |
Hb_001500_120 |
0.1154013771 |
- |
- |
ABC transporter family protein [Hevea brasiliensis] |
| 20 |
Hb_000089_210 |
0.116280123 |
- |
- |
unknown [Medicago truncatula] |