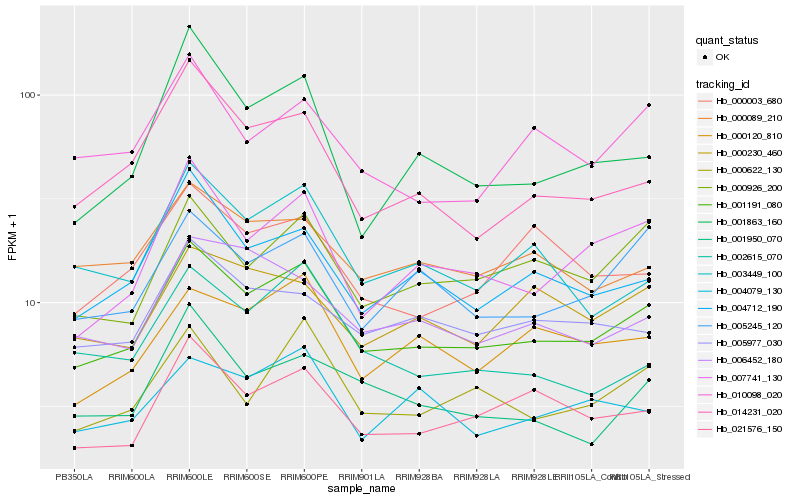
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001191_080 |
0.0 |
- |
- |
protein with unknown function [Ricinus communis] |
| 2 |
Hb_007741_130 |
0.0868308444 |
- |
- |
hypothetical protein B456_007G089000 [Gossypium raimondii] |
| 3 |
Hb_004712_190 |
0.0871199484 |
- |
- |
PREDICTED: uncharacterized protein LOC105633105 [Jatropha curcas] |
| 4 |
Hb_021576_150 |
0.0910055486 |
- |
- |
PREDICTED: formin-like protein 18 [Jatropha curcas] |
| 5 |
Hb_000926_200 |
0.0940962415 |
transcription factor |
TF Family: Rcd1-like |
PREDICTED: cell differentiation protein RCD1 homolog isoform X1 [Jatropha curcas] |
| 6 |
Hb_014231_020 |
0.0973969317 |
- |
- |
PREDICTED: ras-related protein Rab11D [Jatropha curcas] |
| 7 |
Hb_005245_120 |
0.0999014853 |
- |
- |
XPA-binding protein, putative [Ricinus communis] |
| 8 |
Hb_000622_130 |
0.1012740614 |
- |
- |
PREDICTED: tubulin gamma-1 chain [Vitis vinifera] |
| 9 |
Hb_000120_810 |
0.1017620494 |
- |
- |
serine/threonine-protein kinase cx32, putative [Ricinus communis] |
| 10 |
Hb_000089_210 |
0.1018493641 |
- |
- |
unknown [Medicago truncatula] |
| 11 |
Hb_005977_030 |
0.1023171233 |
- |
- |
PREDICTED: monothiol glutaredoxin-S10-like [Jatropha curcas] |
| 12 |
Hb_003449_100 |
0.1043161202 |
- |
- |
PREDICTED: protein YIPF6 homolog [Jatropha curcas] |
| 13 |
Hb_006452_180 |
0.1053977444 |
- |
- |
PREDICTED: BRCA1-associated protein [Jatropha curcas] |
| 14 |
Hb_010098_020 |
0.1068904694 |
- |
- |
PREDICTED: protein BOLA4, chloroplastic/mitochondrial [Jatropha curcas] |
| 15 |
Hb_001950_070 |
0.1077068969 |
- |
- |
cyclin d, putative [Ricinus communis] |
| 16 |
Hb_001863_160 |
0.1100637827 |
- |
- |
PREDICTED: 14-3-3-like protein [Jatropha curcas] |
| 17 |
Hb_002615_070 |
0.1104457804 |
- |
- |
PREDICTED: uncharacterized protein LOC105628992 isoform X1 [Jatropha curcas] |
| 18 |
Hb_004079_130 |
0.1111142614 |
- |
- |
PREDICTED: uncharacterized protein LOC105637708 [Jatropha curcas] |
| 19 |
Hb_000230_460 |
0.1114050639 |
- |
- |
Speckle-type POZ protein, putative [Ricinus communis] |
| 20 |
Hb_000003_680 |
0.1115080617 |
transcription factor |
TF Family: MYB-related |
PREDICTED: myb-like protein H [Jatropha curcas] |