| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
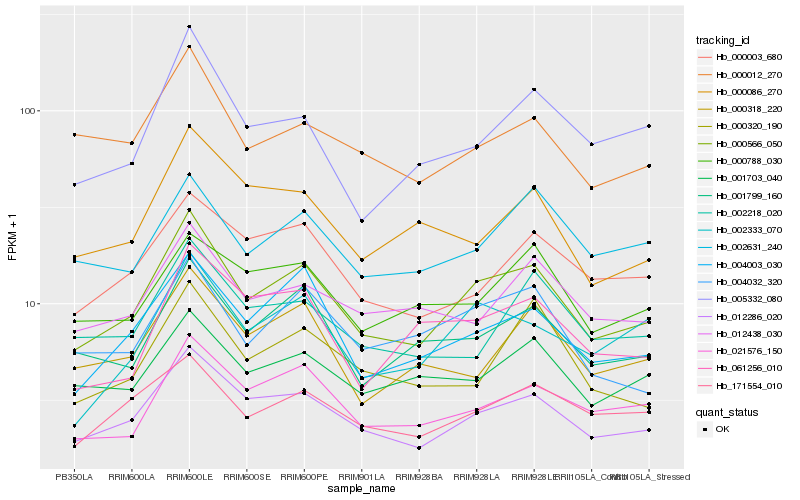
Hb_000566_050 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105644845 [Jatropha curcas] |
| 2 |
Hb_012286_020 |
0.0569145861 |
- |
- |
PREDICTED: uncharacterized protein LOC105648960 [Jatropha curcas] |
| 3 |
Hb_004003_030 |
0.0917391787 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_021576_150 |
0.0953529088 |
- |
- |
PREDICTED: formin-like protein 18 [Jatropha curcas] |
| 5 |
Hb_001799_160 |
0.097099204 |
- |
- |
PREDICTED: protein VASCULAR ASSOCIATED DEATH 1, chloroplastic isoform X2 [Jatropha curcas] |
| 6 |
Hb_171554_010 |
0.1003227124 |
- |
- |
PREDICTED: uncharacterized protein LOC105634071 [Jatropha curcas] |
| 7 |
Hb_004032_320 |
0.1050530787 |
- |
- |
PREDICTED: uncharacterized protein LOC105635153 [Jatropha curcas] |
| 8 |
Hb_002333_070 |
0.1054381271 |
- |
- |
PREDICTED: ACT domain-containing protein ACR4-like [Jatropha curcas] |
| 9 |
Hb_000003_680 |
0.1063688252 |
transcription factor |
TF Family: MYB-related |
PREDICTED: myb-like protein H [Jatropha curcas] |
| 10 |
Hb_005332_080 |
0.1081836046 |
- |
- |
PREDICTED: succinate dehydrogenase [ubiquinone] iron-sulfur subunit 2, mitochondrial [Jatropha curcas] |
| 11 |
Hb_000012_270 |
0.1083198474 |
- |
- |
NADH-plastoquinone oxidoreductase, putative [Ricinus communis] |
| 12 |
Hb_001703_040 |
0.109330375 |
- |
- |
PREDICTED: uncharacterized protein At1g04910 isoform X2 [Jatropha curcas] |
| 13 |
Hb_000318_220 |
0.1101809639 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_012438_030 |
0.1110159877 |
- |
- |
PREDICTED: protein sym-1 [Jatropha curcas] |
| 15 |
Hb_002631_240 |
0.1121987994 |
- |
- |
JHL17M24.3 [Jatropha curcas] |
| 16 |
Hb_002218_020 |
0.1140697321 |
- |
- |
PREDICTED: AT-hook motif nuclear-localized protein 14 [Jatropha curcas] |
| 17 |
Hb_000086_270 |
0.1150953006 |
- |
- |
hypothetical protein POPTR_0002s03730g [Populus trichocarpa] |
| 18 |
Hb_000788_030 |
0.1154836978 |
- |
- |
PREDICTED: dynamin-2A [Jatropha curcas] |
| 19 |
Hb_061256_010 |
0.1167142231 |
- |
- |
PREDICTED: uncharacterized protein LOC105635220 isoform X2 [Jatropha curcas] |
| 20 |
Hb_000320_190 |
0.1178983874 |
- |
- |
PREDICTED: aminomethyltransferase, mitochondrial [Jatropha curcas] |