| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
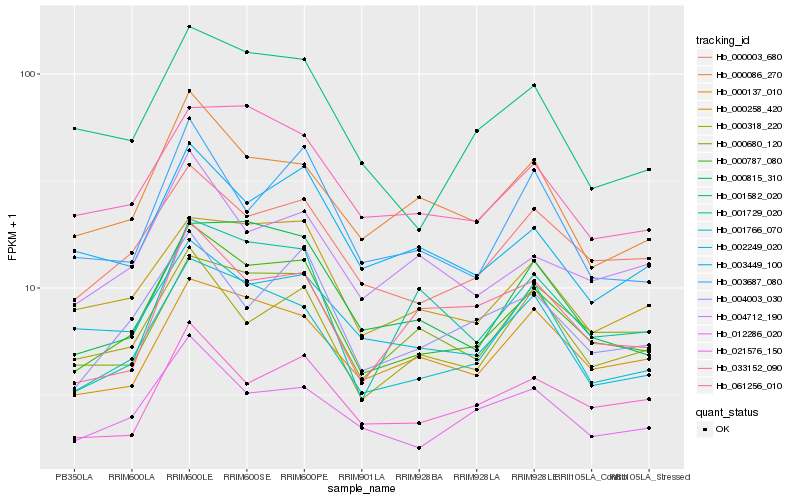
Hb_000787_080 |
0.0 |
- |
- |
PREDICTED: TBC1 domain family member 16-like [Jatropha curcas] |
| 2 |
Hb_001766_070 |
0.0796005017 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_000003_680 |
0.0796383591 |
transcription factor |
TF Family: MYB-related |
PREDICTED: myb-like protein H [Jatropha curcas] |
| 4 |
Hb_000258_420 |
0.0860269423 |
- |
- |
sucrose transporter 2A [Hevea brasiliensis] |
| 5 |
Hb_000815_310 |
0.0914470209 |
- |
- |
protein kinase, putative [Ricinus communis] |
| 6 |
Hb_033152_090 |
0.0923879211 |
- |
- |
PREDICTED: serine/threonine-protein kinase SRK2E isoform X2 [Gossypium raimondii] |
| 7 |
Hb_000137_010 |
0.0927863973 |
- |
- |
PREDICTED: uncharacterized protein LOC105644349 [Jatropha curcas] |
| 8 |
Hb_000680_120 |
0.0935208405 |
- |
- |
PREDICTED: uncharacterized protein LOC105631461 [Jatropha curcas] |
| 9 |
Hb_004003_030 |
0.0948953123 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_000318_220 |
0.0953288119 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_000086_270 |
0.0962891089 |
- |
- |
hypothetical protein POPTR_0002s03730g [Populus trichocarpa] |
| 12 |
Hb_021576_150 |
0.0965396694 |
- |
- |
PREDICTED: formin-like protein 18 [Jatropha curcas] |
| 13 |
Hb_012286_020 |
0.0968758909 |
- |
- |
PREDICTED: uncharacterized protein LOC105648960 [Jatropha curcas] |
| 14 |
Hb_003449_100 |
0.1017873846 |
- |
- |
PREDICTED: protein YIPF6 homolog [Jatropha curcas] |
| 15 |
Hb_004712_190 |
0.1035484739 |
- |
- |
PREDICTED: uncharacterized protein LOC105633105 [Jatropha curcas] |
| 16 |
Hb_001582_020 |
0.1035729541 |
- |
- |
plastid acyl-ACP thioesterase [Vernicia fordii] |
| 17 |
Hb_061256_010 |
0.1059347597 |
- |
- |
PREDICTED: uncharacterized protein LOC105635220 isoform X2 [Jatropha curcas] |
| 18 |
Hb_002249_020 |
0.1061566493 |
- |
- |
PREDICTED: dual specificity protein phosphatase PHS1 [Jatropha curcas] |
| 19 |
Hb_003687_080 |
0.1079777295 |
- |
- |
PREDICTED: casein kinase I-like [Jatropha curcas] |
| 20 |
Hb_001729_020 |
0.1099914792 |
transcription factor |
TF Family: C3H |
zinc finger protein, putative [Ricinus communis] |