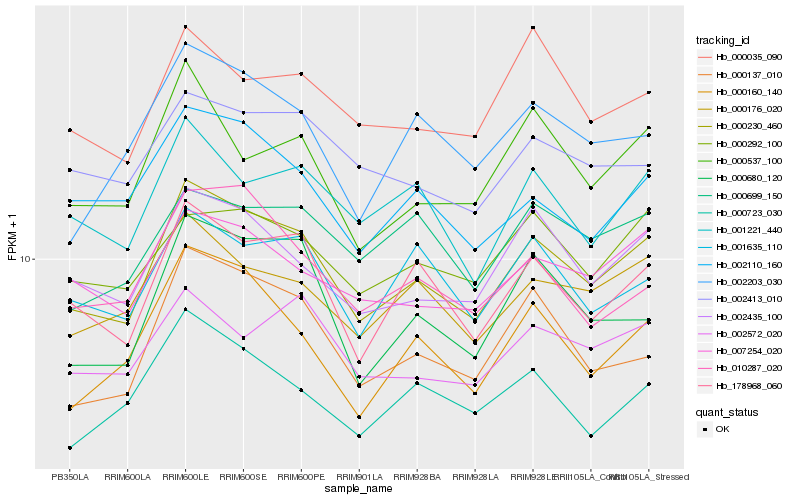
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000230_460 |
0.0 |
- |
- |
Speckle-type POZ protein, putative [Ricinus communis] |
| 2 |
Hb_178968_060 |
0.0631502841 |
- |
- |
PREDICTED: ubiquitin receptor RAD23b-like [Gossypium raimondii] |
| 3 |
Hb_002435_100 |
0.0690887943 |
- |
- |
PREDICTED: probable E3 ubiquitin-protein ligase RNF217 [Jatropha curcas] |
| 4 |
Hb_000176_020 |
0.0725363621 |
- |
- |
PREDICTED: nuclear pore complex protein NUP43 [Jatropha curcas] |
| 5 |
Hb_000137_010 |
0.0778991534 |
- |
- |
PREDICTED: uncharacterized protein LOC105644349 [Jatropha curcas] |
| 6 |
Hb_007254_020 |
0.0794881018 |
- |
- |
PREDICTED: uncharacterized protein LOC105629137 [Jatropha curcas] |
| 7 |
Hb_000160_140 |
0.0795328723 |
- |
- |
PREDICTED: ELL-associated factor 2-like isoform X1 [Jatropha curcas] |
| 8 |
Hb_000680_120 |
0.0800133686 |
- |
- |
PREDICTED: uncharacterized protein LOC105631461 [Jatropha curcas] |
| 9 |
Hb_001221_440 |
0.0803518115 |
- |
- |
PREDICTED: SUMO-activating enzyme subunit 1B-1 isoform X1 [Jatropha curcas] |
| 10 |
Hb_001635_110 |
0.081696319 |
- |
- |
PREDICTED: NADP-specific glutamate dehydrogenase [Jatropha curcas] |
| 11 |
Hb_002110_160 |
0.0825134239 |
- |
- |
PREDICTED: phosphoribosylaminoimidazole-succinocarboxamide synthase, chloroplastic [Jatropha curcas] |
| 12 |
Hb_000537_100 |
0.0826793239 |
- |
- |
Coiled-coil domain-containing protein, putative [Ricinus communis] |
| 13 |
Hb_000292_100 |
0.0829295465 |
transcription factor |
TF Family: bZIP |
PREDICTED: transcription factor HBP-1a [Jatropha curcas] |
| 14 |
Hb_010287_020 |
0.0842943326 |
- |
- |
CCR4-NOT transcription complex subunit, putative [Ricinus communis] |
| 15 |
Hb_002572_020 |
0.0842989351 |
transcription factor |
TF Family: SBP |
hypothetical protein POPTR_0005s28010g [Populus trichocarpa] |
| 16 |
Hb_000723_030 |
0.0860169918 |
transcription factor |
TF Family: E2F-DP |
PREDICTED: transcription factor-like protein DPB isoform X1 [Jatropha curcas] |
| 17 |
Hb_002203_030 |
0.0862717288 |
transcription factor |
TF Family: NF-YB |
PREDICTED: protein Dr1 homolog isoform X2 [Jatropha curcas] |
| 18 |
Hb_002413_010 |
0.0879395524 |
- |
- |
ABC transporter family protein [Hevea brasiliensis] |
| 19 |
Hb_000699_150 |
0.0879455113 |
- |
- |
PREDICTED: AT-hook motif nuclear-localized protein 14 [Jatropha curcas] |
| 20 |
Hb_000035_090 |
0.088030246 |
- |
- |
PREDICTED: stromal cell-derived factor 2-like protein [Jatropha curcas] |