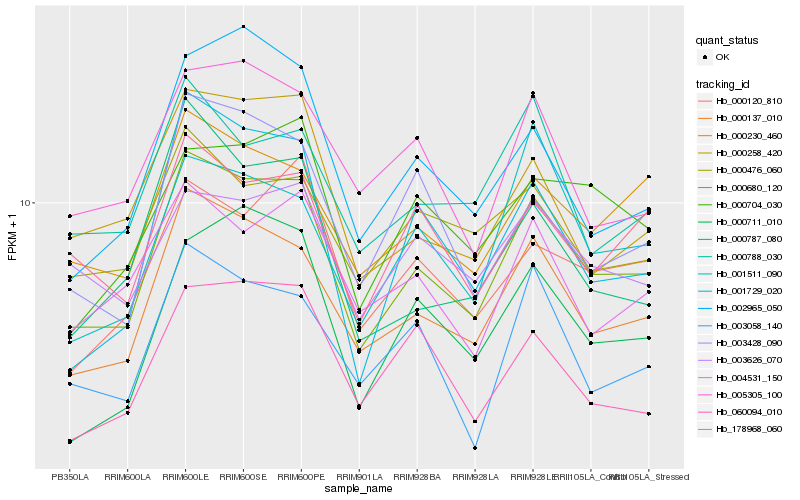
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000680_120 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105631461 [Jatropha curcas] |
| 2 |
Hb_000137_010 |
0.0485169341 |
- |
- |
PREDICTED: uncharacterized protein LOC105644349 [Jatropha curcas] |
| 3 |
Hb_000230_460 |
0.0800133686 |
- |
- |
Speckle-type POZ protein, putative [Ricinus communis] |
| 4 |
Hb_000258_420 |
0.0844415567 |
- |
- |
sucrose transporter 2A [Hevea brasiliensis] |
| 5 |
Hb_000120_810 |
0.0868661083 |
- |
- |
serine/threonine-protein kinase cx32, putative [Ricinus communis] |
| 6 |
Hb_178968_060 |
0.0873372448 |
- |
- |
PREDICTED: ubiquitin receptor RAD23b-like [Gossypium raimondii] |
| 7 |
Hb_000788_030 |
0.0909039942 |
- |
- |
PREDICTED: dynamin-2A [Jatropha curcas] |
| 8 |
Hb_003428_090 |
0.0912016639 |
- |
- |
PREDICTED: DNA (cytosine-5)-methyltransferase DRM2 [Jatropha curcas] |
| 9 |
Hb_005305_100 |
0.0912739396 |
transcription factor |
TF Family: NF-YA |
PREDICTED: nuclear transcription factor Y subunit A-1-like isoform X1 [Jatropha curcas] |
| 10 |
Hb_004531_150 |
0.0914248273 |
- |
- |
PREDICTED: peroxisome biogenesis protein 2 isoform X1 [Jatropha curcas] |
| 11 |
Hb_000711_010 |
0.0915558897 |
transcription factor |
TF Family: Tify |
PREDICTED: protein TIFY 6B isoform X1 [Jatropha curcas] |
| 12 |
Hb_003058_140 |
0.0917562276 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_000704_030 |
0.0919602599 |
- |
- |
PREDICTED: probable aquaporin SIP2-1 [Cicer arietinum] |
| 14 |
Hb_001511_090 |
0.0928574971 |
- |
- |
PREDICTED: uncharacterized protein LOC105645861 [Jatropha curcas] |
| 15 |
Hb_000476_060 |
0.0929809655 |
- |
- |
Tetratricopeptide repeat (TPR)-like superfamily protein isoform 1 [Theobroma cacao] |
| 16 |
Hb_000787_080 |
0.0935208405 |
- |
- |
PREDICTED: TBC1 domain family member 16-like [Jatropha curcas] |
| 17 |
Hb_003626_070 |
0.0937584479 |
- |
- |
PREDICTED: golgin candidate 1 [Jatropha curcas] |
| 18 |
Hb_060094_010 |
0.0941014443 |
- |
- |
PREDICTED: C2 and GRAM domain-containing protein At1g03370 isoform X2 [Jatropha curcas] |
| 19 |
Hb_002965_050 |
0.0941362246 |
transcription factor |
TF Family: ARR-B |
PREDICTED: two-component response regulator ARR2 [Jatropha curcas] |
| 20 |
Hb_001729_020 |
0.0949263015 |
transcription factor |
TF Family: C3H |
zinc finger protein, putative [Ricinus communis] |