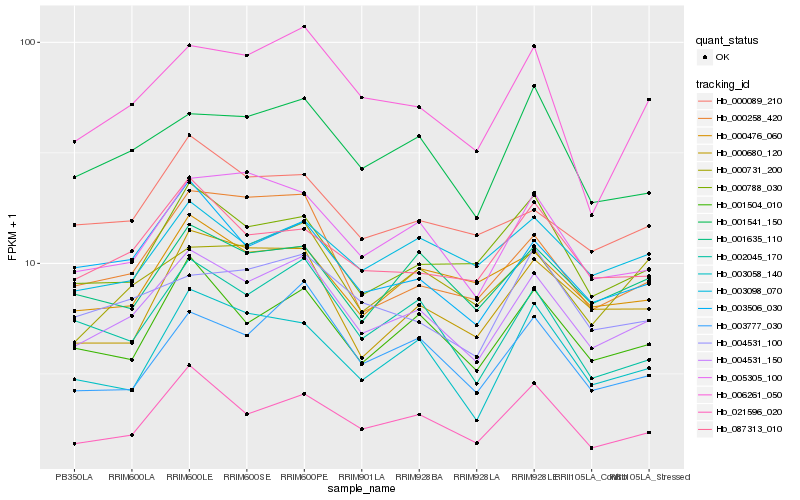
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004531_150 |
0.0 |
- |
- |
PREDICTED: peroxisome biogenesis protein 2 isoform X1 [Jatropha curcas] |
| 2 |
Hb_001504_010 |
0.0754313035 |
- |
- |
PREDICTED: uncharacterized protein LOC105645377 [Jatropha curcas] |
| 3 |
Hb_005305_100 |
0.0784306557 |
transcription factor |
TF Family: NF-YA |
PREDICTED: nuclear transcription factor Y subunit A-1-like isoform X1 [Jatropha curcas] |
| 4 |
Hb_003506_030 |
0.0793584179 |
- |
- |
PREDICTED: ATP-dependent Clp protease ATP-binding subunit clpX-like, mitochondrial isoform X2 [Jatropha curcas] |
| 5 |
Hb_006261_050 |
0.0815830731 |
transcription factor |
TF Family: C2C2-YABBY |
DNA-binding protein MNB1B, putative [Ricinus communis] |
| 6 |
Hb_001541_150 |
0.081688905 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_021596_020 |
0.0819821656 |
- |
- |
hypothetical protein JCGZ_02034 [Jatropha curcas] |
| 8 |
Hb_002045_170 |
0.08482915 |
- |
- |
interferon-induced guanylate-binding protein, putative [Ricinus communis] |
| 9 |
Hb_000258_420 |
0.0854484746 |
- |
- |
sucrose transporter 2A [Hevea brasiliensis] |
| 10 |
Hb_003777_030 |
0.0859313273 |
- |
- |
Exocyst complex component, putative [Ricinus communis] |
| 11 |
Hb_000788_030 |
0.0865704299 |
- |
- |
PREDICTED: dynamin-2A [Jatropha curcas] |
| 12 |
Hb_087313_010 |
0.087120008 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_001635_110 |
0.0877607823 |
- |
- |
PREDICTED: NADP-specific glutamate dehydrogenase [Jatropha curcas] |
| 14 |
Hb_000731_200 |
0.0888801652 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g61870, mitochondrial [Jatropha curcas] |
| 15 |
Hb_000476_060 |
0.0903735998 |
- |
- |
Tetratricopeptide repeat (TPR)-like superfamily protein isoform 1 [Theobroma cacao] |
| 16 |
Hb_000089_210 |
0.0910739272 |
- |
- |
unknown [Medicago truncatula] |
| 17 |
Hb_000680_120 |
0.0914248273 |
- |
- |
PREDICTED: uncharacterized protein LOC105631461 [Jatropha curcas] |
| 18 |
Hb_003058_140 |
0.09148807 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_003098_070 |
0.092282022 |
- |
- |
PREDICTED: uncharacterized protein LOC105633456 [Jatropha curcas] |
| 20 |
Hb_004531_100 |
0.0924492904 |
- |
- |
PREDICTED: zinc phosphodiesterase ELAC protein 2 isoform X1 [Jatropha curcas] |