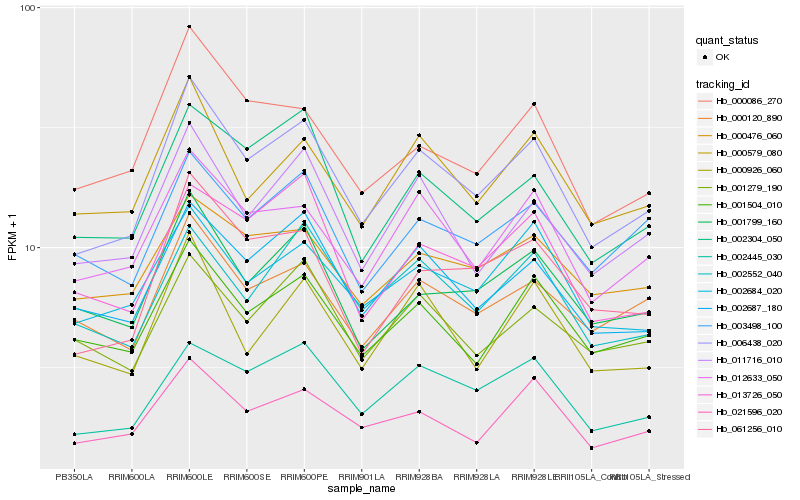
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_006438_020 |
0.0 |
- |
- |
PREDICTED: lysine--tRNA ligase-like [Populus euphratica] |
| 2 |
Hb_012633_050 |
0.0707713695 |
- |
- |
zinc finger protein, putative [Ricinus communis] |
| 3 |
Hb_002445_030 |
0.0727336309 |
- |
- |
hypothetical protein JCGZ_12656 [Jatropha curcas] |
| 4 |
Hb_002687_180 |
0.074152861 |
- |
- |
PREDICTED: phytochrome-associated serine/threonine-protein phosphatase [Jatropha curcas] |
| 5 |
Hb_011716_010 |
0.0764210403 |
- |
- |
PREDICTED: probable protein arginine N-methyltransferase 1.2 [Jatropha curcas] |
| 6 |
Hb_021596_020 |
0.0770037922 |
- |
- |
hypothetical protein JCGZ_02034 [Jatropha curcas] |
| 7 |
Hb_061256_010 |
0.0787981335 |
- |
- |
PREDICTED: uncharacterized protein LOC105635220 isoform X2 [Jatropha curcas] |
| 8 |
Hb_000579_080 |
0.080109445 |
- |
- |
PREDICTED: 26S protease regulatory subunit S10B homolog B [Jatropha curcas] |
| 9 |
Hb_001504_010 |
0.080849053 |
- |
- |
PREDICTED: uncharacterized protein LOC105645377 [Jatropha curcas] |
| 10 |
Hb_002304_050 |
0.0810763619 |
- |
- |
pyruvate kinase, putative [Ricinus communis] |
| 11 |
Hb_000120_890 |
0.084123415 |
- |
- |
PREDICTED: phosphoinositide phosphatase SAC6 [Jatropha curcas] |
| 12 |
Hb_001799_160 |
0.0847875385 |
- |
- |
PREDICTED: protein VASCULAR ASSOCIATED DEATH 1, chloroplastic isoform X2 [Jatropha curcas] |
| 13 |
Hb_000086_270 |
0.0855730552 |
- |
- |
hypothetical protein POPTR_0002s03730g [Populus trichocarpa] |
| 14 |
Hb_002684_020 |
0.0860069641 |
- |
- |
ATP-dependent clp protease ATP-binding subunit clpx, putative [Ricinus communis] |
| 15 |
Hb_002552_040 |
0.0887680122 |
- |
- |
PREDICTED: uncharacterized protein LOC105641220 [Jatropha curcas] |
| 16 |
Hb_000476_060 |
0.090371723 |
- |
- |
Tetratricopeptide repeat (TPR)-like superfamily protein isoform 1 [Theobroma cacao] |
| 17 |
Hb_013726_050 |
0.0923765768 |
- |
- |
PREDICTED: probable methyltransferase PMT13 [Jatropha curcas] |
| 18 |
Hb_003498_100 |
0.0926794855 |
- |
- |
component of oligomeric golgi complex, putative [Ricinus communis] |
| 19 |
Hb_001279_190 |
0.09300612 |
- |
- |
PREDICTED: uncharacterized membrane protein At3g27390 isoform X1 [Jatropha curcas] |
| 20 |
Hb_000926_060 |
0.0942711163 |
- |
- |
conserved hypothetical protein [Ricinus communis] |