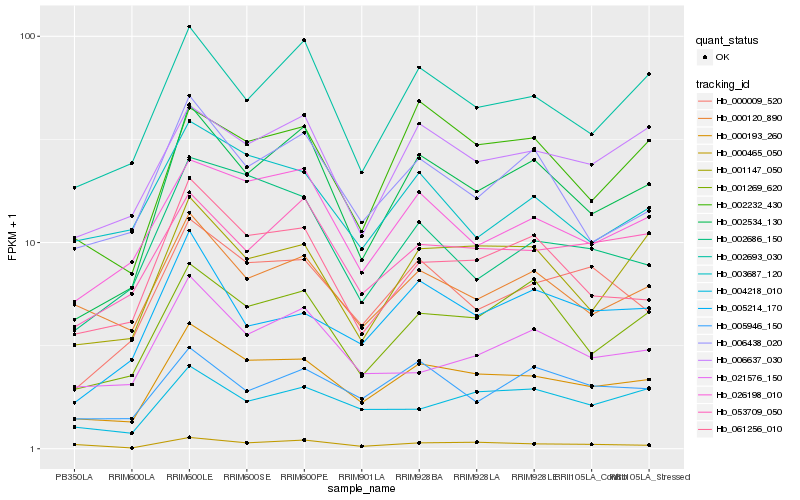
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000193_260 |
0.0 |
- |
- |
PREDICTED: protein NEN1 [Jatropha curcas] |
| 2 |
Hb_002534_130 |
0.0876113371 |
- |
- |
membrane associated ring finger 1,8, putative [Ricinus communis] |
| 3 |
Hb_061256_010 |
0.0954672178 |
- |
- |
PREDICTED: uncharacterized protein LOC105635220 isoform X2 [Jatropha curcas] |
| 4 |
Hb_026198_010 |
0.096616344 |
- |
- |
PREDICTED: uncharacterized protein LOC105634369 isoform X1 [Jatropha curcas] |
| 5 |
Hb_000009_520 |
0.0980490549 |
- |
- |
PREDICTED: wall-associated receptor kinase-like 14 [Jatropha curcas] |
| 6 |
Hb_002693_030 |
0.0997430451 |
- |
- |
PREDICTED: mitochondrial outer membrane protein porin of 34 kDa [Jatropha curcas] |
| 7 |
Hb_001147_050 |
0.1009699385 |
- |
- |
PREDICTED: probable serine/threonine-protein kinase Cx32, chloroplastic [Jatropha curcas] |
| 8 |
Hb_006637_030 |
0.1043856214 |
- |
- |
hypothetical protein CISIN_1g017413mg [Citrus sinensis] |
| 9 |
Hb_001269_620 |
0.1044230829 |
- |
- |
PREDICTED: magnesium transporter MRS2-1 isoform X1 [Populus euphratica] |
| 10 |
Hb_000120_890 |
0.1047374149 |
- |
- |
PREDICTED: phosphoinositide phosphatase SAC6 [Jatropha curcas] |
| 11 |
Hb_002232_430 |
0.1076871262 |
- |
- |
PREDICTED: soluble inorganic pyrophosphatase 6, chloroplastic [Jatropha curcas] |
| 12 |
Hb_053709_050 |
0.1081854986 |
- |
- |
PREDICTED: nudix hydrolase 23, chloroplastic isoform X2 [Jatropha curcas] |
| 13 |
Hb_006438_020 |
0.1101245854 |
- |
- |
PREDICTED: lysine--tRNA ligase-like [Populus euphratica] |
| 14 |
Hb_004218_010 |
0.1113136648 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: putative disease resistance RPP13-like protein 1 [Jatropha curcas] |
| 15 |
Hb_000465_050 |
0.1141545643 |
- |
- |
PREDICTED: DNA mismatch repair protein MSH4 [Jatropha curcas] |
| 16 |
Hb_005946_150 |
0.1159304588 |
- |
- |
PREDICTED: DNA mismatch repair protein MSH7 [Jatropha curcas] |
| 17 |
Hb_005214_170 |
0.116573093 |
- |
- |
PREDICTED: uncharacterized protein LOC105636021 [Jatropha curcas] |
| 18 |
Hb_003687_120 |
0.1166840826 |
- |
- |
PREDICTED: serine/threonine-protein kinase tricorner-like [Jatropha curcas] |
| 19 |
Hb_002686_150 |
0.1170618006 |
- |
- |
Glucan endo-1,3-beta-glucosidase precursor, putative [Ricinus communis] |
| 20 |
Hb_021576_150 |
0.1175506923 |
- |
- |
PREDICTED: formin-like protein 18 [Jatropha curcas] |