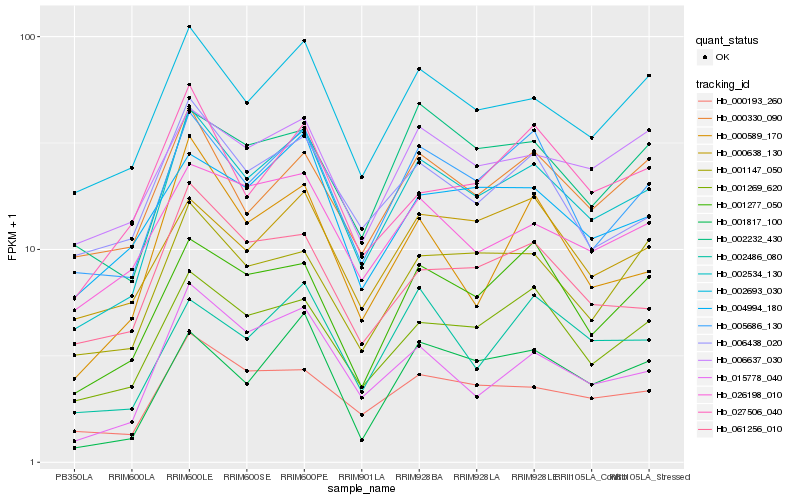
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002534_130 |
0.0 |
- |
- |
membrane associated ring finger 1,8, putative [Ricinus communis] |
| 2 |
Hb_002693_030 |
0.0827480966 |
- |
- |
PREDICTED: mitochondrial outer membrane protein porin of 34 kDa [Jatropha curcas] |
| 3 |
Hb_005686_130 |
0.0827696416 |
rubber biosynthesis |
Gene Name: Isopentenyl-diphosphate delta isomerase |
isopentenyl pyrophosphate isomerase [Hevea brasiliensis] |
| 4 |
Hb_000193_260 |
0.0876113371 |
- |
- |
PREDICTED: protein NEN1 [Jatropha curcas] |
| 5 |
Hb_001269_620 |
0.0889897692 |
- |
- |
PREDICTED: magnesium transporter MRS2-1 isoform X1 [Populus euphratica] |
| 6 |
Hb_015778_040 |
0.092878855 |
- |
- |
PREDICTED: uncharacterized protein LOC105650695 [Jatropha curcas] |
| 7 |
Hb_001277_050 |
0.1036022351 |
- |
- |
PREDICTED: pyruvate kinase isozyme A, chloroplastic [Jatropha curcas] |
| 8 |
Hb_006438_020 |
0.1070497308 |
- |
- |
PREDICTED: lysine--tRNA ligase-like [Populus euphratica] |
| 9 |
Hb_061256_010 |
0.1080519778 |
- |
- |
PREDICTED: uncharacterized protein LOC105635220 isoform X2 [Jatropha curcas] |
| 10 |
Hb_026198_010 |
0.1084281911 |
- |
- |
PREDICTED: uncharacterized protein LOC105634369 isoform X1 [Jatropha curcas] |
| 11 |
Hb_001147_050 |
0.1097957667 |
- |
- |
PREDICTED: probable serine/threonine-protein kinase Cx32, chloroplastic [Jatropha curcas] |
| 12 |
Hb_000330_090 |
0.1105042878 |
- |
- |
ornithine carbamoyltransferase, putative [Ricinus communis] |
| 13 |
Hb_006637_030 |
0.1131737409 |
- |
- |
hypothetical protein CISIN_1g017413mg [Citrus sinensis] |
| 14 |
Hb_001817_100 |
0.1132189829 |
- |
- |
PREDICTED: deoxyhypusine hydroxylase [Jatropha curcas] |
| 15 |
Hb_004994_180 |
0.1132894155 |
- |
- |
malic enzyme, putative [Ricinus communis] |
| 16 |
Hb_027506_040 |
0.1136772898 |
rubber biosynthesis |
Gene Name: Pyruvate dehydrogenase |
PREDICTED: pyruvate dehydrogenase E1 component subunit beta-3, chloroplastic [Jatropha curcas] |
| 17 |
Hb_000589_170 |
0.1141888157 |
- |
- |
PREDICTED: telomere repeat-binding protein 4 isoform X3 [Jatropha curcas] |
| 18 |
Hb_002232_430 |
0.1144780973 |
- |
- |
PREDICTED: soluble inorganic pyrophosphatase 6, chloroplastic [Jatropha curcas] |
| 19 |
Hb_000638_130 |
0.1160382419 |
- |
- |
PREDICTED: SAL1 phosphatase-like [Jatropha curcas] |
| 20 |
Hb_002486_080 |
0.1165337357 |
- |
- |
PREDICTED: cytosolic endo-beta-N-acetylglucosaminidase isoform X1 [Jatropha curcas] |