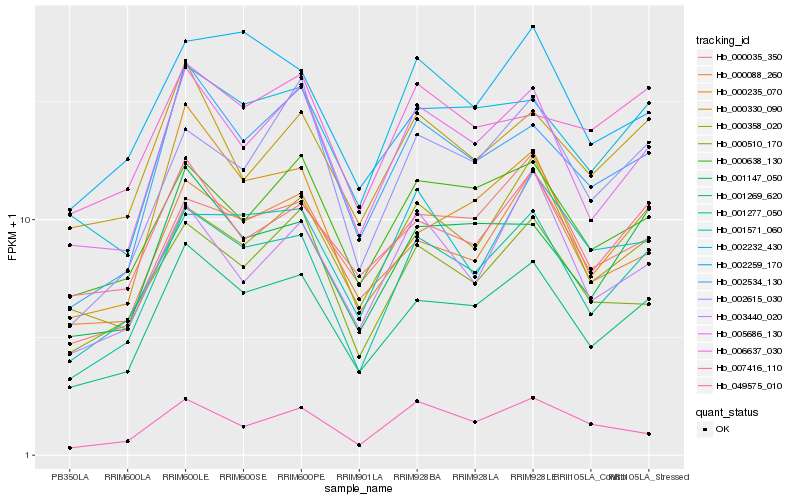
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001277_050 |
0.0 |
- |
- |
PREDICTED: pyruvate kinase isozyme A, chloroplastic [Jatropha curcas] |
| 2 |
Hb_001269_620 |
0.0601591736 |
- |
- |
PREDICTED: magnesium transporter MRS2-1 isoform X1 [Populus euphratica] |
| 3 |
Hb_005686_130 |
0.0815866376 |
rubber biosynthesis |
Gene Name: Isopentenyl-diphosphate delta isomerase |
isopentenyl pyrophosphate isomerase [Hevea brasiliensis] |
| 4 |
Hb_001147_050 |
0.0984387065 |
- |
- |
PREDICTED: probable serine/threonine-protein kinase Cx32, chloroplastic [Jatropha curcas] |
| 5 |
Hb_000088_260 |
0.1007826366 |
- |
- |
PREDICTED: uncharacterized protein LOC105636987 [Jatropha curcas] |
| 6 |
Hb_000638_130 |
0.1032215564 |
- |
- |
PREDICTED: SAL1 phosphatase-like [Jatropha curcas] |
| 7 |
Hb_002534_130 |
0.1036022351 |
- |
- |
membrane associated ring finger 1,8, putative [Ricinus communis] |
| 8 |
Hb_000035_350 |
0.1046058954 |
- |
- |
PREDICTED: monogalactosyldiacylglycerol synthase, chloroplastic [Jatropha curcas] |
| 9 |
Hb_001571_060 |
0.1049121642 |
- |
- |
PREDICTED: uncharacterized protein LOC105643976 [Jatropha curcas] |
| 10 |
Hb_002232_430 |
0.1062516652 |
- |
- |
PREDICTED: soluble inorganic pyrophosphatase 6, chloroplastic [Jatropha curcas] |
| 11 |
Hb_000358_020 |
0.1086395427 |
- |
- |
PREDICTED: uncharacterized protein C05D11.7-like [Jatropha curcas] |
| 12 |
Hb_000510_170 |
0.109668398 |
- |
- |
Glycosyltransferase QUASIMODO1, putative [Ricinus communis] |
| 13 |
Hb_049575_010 |
0.1109930216 |
- |
- |
hypothetical protein POPTR_0001s15330g [Populus trichocarpa] |
| 14 |
Hb_002615_030 |
0.1116628783 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RHF1A-like [Populus euphratica] |
| 15 |
Hb_003440_020 |
0.1118309089 |
- |
- |
PREDICTED: serine protease SPPA, chloroplastic isoform X2 [Jatropha curcas] |
| 16 |
Hb_000330_090 |
0.1163051108 |
- |
- |
ornithine carbamoyltransferase, putative [Ricinus communis] |
| 17 |
Hb_002259_170 |
0.1166898727 |
- |
- |
PREDICTED: internal alternative NAD(P)H-ubiquinone oxidoreductase A1, mitochondrial-like [Jatropha curcas] |
| 18 |
Hb_006637_030 |
0.117137381 |
- |
- |
hypothetical protein CISIN_1g017413mg [Citrus sinensis] |
| 19 |
Hb_007416_110 |
0.1182408818 |
- |
- |
PREDICTED: probable cyclic nucleotide-gated ion channel 14 isoform X1 [Jatropha curcas] |
| 20 |
Hb_000235_070 |
0.1192529065 |
- |
- |
PREDICTED: VIN3-like protein 2 [Jatropha curcas] |