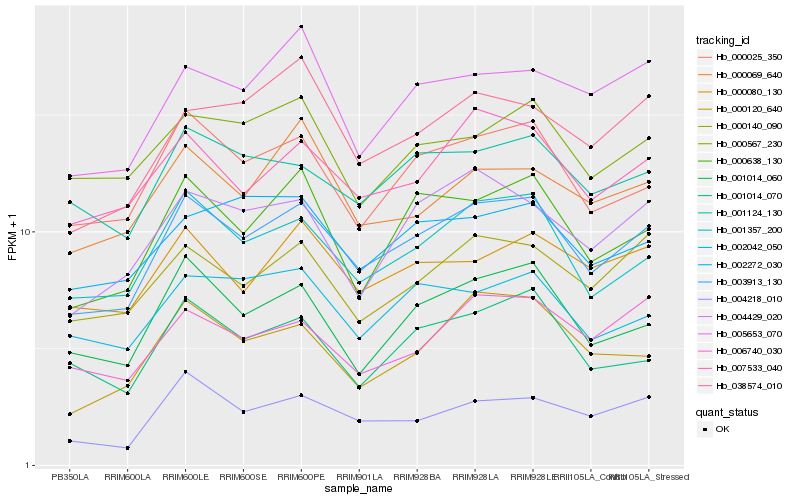
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003913_130 |
0.0 |
- |
- |
histone deacetylase 1, 2 ,3, putative [Ricinus communis] |
| 2 |
Hb_001357_200 |
0.0612449012 |
transcription factor |
TF Family: Rcd1-like |
PREDICTED: cell differentiation protein RCD1 homolog isoform X1 [Jatropha curcas] |
| 3 |
Hb_000025_350 |
0.0681127516 |
- |
- |
PREDICTED: rho GTPase-activating protein 7 isoform X1 [Jatropha curcas] |
| 4 |
Hb_007533_040 |
0.074192271 |
- |
- |
protein phosphatase 2c, putative [Ricinus communis] |
| 5 |
Hb_000638_130 |
0.0787040828 |
- |
- |
PREDICTED: SAL1 phosphatase-like [Jatropha curcas] |
| 6 |
Hb_000140_090 |
0.0799305143 |
- |
- |
PREDICTED: C-terminal binding protein AN [Jatropha curcas] |
| 7 |
Hb_001014_060 |
0.0800867835 |
transcription factor |
TF Family: SET |
histone-lysine n-methyltransferase, suvh, putative [Ricinus communis] |
| 8 |
Hb_004429_020 |
0.0841561311 |
- |
- |
PREDICTED: magnesium transporter MRS2-5 [Jatropha curcas] |
| 9 |
Hb_001124_130 |
0.0849628111 |
transcription factor |
TF Family: TUB |
phosphoric diester hydrolase, putative [Ricinus communis] |
| 10 |
Hb_000080_130 |
0.0869678456 |
- |
- |
PREDICTED: TBC1 domain family member 22B [Jatropha curcas] |
| 11 |
Hb_004218_010 |
0.0874814979 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: putative disease resistance RPP13-like protein 1 [Jatropha curcas] |
| 12 |
Hb_002042_050 |
0.0887548044 |
- |
- |
PREDICTED: uncharacterized protein LOC105647554 isoform X1 [Jatropha curcas] |
| 13 |
Hb_000120_640 |
0.0894813239 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 14 |
Hb_002272_030 |
0.0907958937 |
- |
- |
PREDICTED: cation/calcium exchanger 5 [Jatropha curcas] |
| 15 |
Hb_005653_070 |
0.0910132949 |
- |
- |
K+ transport growth defect-like protein [Hevea brasiliensis] |
| 16 |
Hb_006740_030 |
0.0917491387 |
- |
- |
PREDICTED: calcium uniporter protein 6, mitochondrial-like [Jatropha curcas] |
| 17 |
Hb_001014_070 |
0.0918136971 |
- |
- |
PREDICTED: uncharacterized protein LOC105647305 [Jatropha curcas] |
| 18 |
Hb_038574_010 |
0.0922830271 |
- |
- |
PREDICTED: vacuolar cation/proton exchanger 3-like isoform X2 [Populus euphratica] |
| 19 |
Hb_000069_640 |
0.0938250641 |
- |
- |
PREDICTED: tubulin gamma-1 chain [Vitis vinifera] |
| 20 |
Hb_000567_230 |
0.093957372 |
- |
- |
PREDICTED: peroxisome biogenesis protein 7 [Jatropha curcas] |