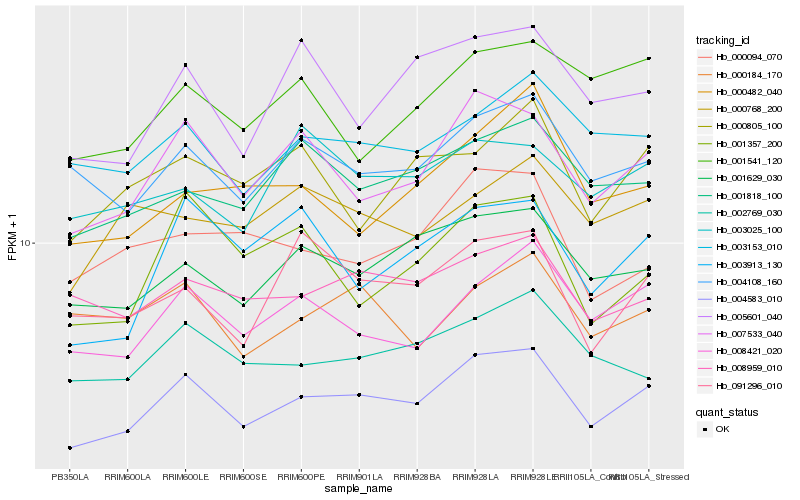
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004583_010 |
0.0 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At3g22150, chloroplastic [Jatropha curcas] |
| 2 |
Hb_007533_040 |
0.0661369106 |
- |
- |
protein phosphatase 2c, putative [Ricinus communis] |
| 3 |
Hb_001629_030 |
0.0785715696 |
- |
- |
PAP-specific phosphatase HAL2-like family protein [Populus trichocarpa] |
| 4 |
Hb_008421_020 |
0.0801752576 |
- |
- |
PREDICTED: uncharacterized protein LOC105635546 isoform X2 [Jatropha curcas] |
| 5 |
Hb_004108_160 |
0.0833096818 |
- |
- |
PREDICTED: myosin heavy chain, non-muscle-like [Jatropha curcas] |
| 6 |
Hb_003153_010 |
0.0886909695 |
- |
- |
PREDICTED: diacylglycerol kinase 3-like [Jatropha curcas] |
| 7 |
Hb_001818_100 |
0.0891383475 |
- |
- |
PREDICTED: DEAD-box ATP-dependent RNA helicase 11-like [Jatropha curcas] |
| 8 |
Hb_002769_030 |
0.0892538168 |
- |
- |
PREDICTED: MATE efflux family protein 4, chloroplastic-like [Jatropha curcas] |
| 9 |
Hb_008959_010 |
0.0928306842 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_005601_040 |
0.0939759351 |
- |
- |
Histone deacetylase 1 isoform 1 [Theobroma cacao] |
| 11 |
Hb_091296_010 |
0.0969711471 |
- |
- |
PREDICTED: uncharacterized protein LOC105124982 [Populus euphratica] |
| 12 |
Hb_003913_130 |
0.0978182708 |
- |
- |
histone deacetylase 1, 2 ,3, putative [Ricinus communis] |
| 13 |
Hb_000805_100 |
0.1008295384 |
- |
- |
PREDICTED: ATP-dependent zinc metalloprotease FTSH 10, mitochondrial isoform X1 [Jatropha curcas] |
| 14 |
Hb_000094_070 |
0.1011213972 |
- |
- |
PREDICTED: 5'-nucleotidase domain-containing protein 4 isoform X2 [Jatropha curcas] |
| 15 |
Hb_001541_120 |
0.1018312324 |
- |
- |
hypothetical protein JCGZ_17842 [Jatropha curcas] |
| 16 |
Hb_000768_200 |
0.1033560723 |
- |
- |
hypothetical protein POPTR_0013s06910g [Populus trichocarpa] |
| 17 |
Hb_000482_040 |
0.1034540123 |
- |
- |
Protein YME1, putative [Ricinus communis] |
| 18 |
Hb_001357_200 |
0.103542887 |
transcription factor |
TF Family: Rcd1-like |
PREDICTED: cell differentiation protein RCD1 homolog isoform X1 [Jatropha curcas] |
| 19 |
Hb_000184_170 |
0.1035793408 |
- |
- |
hypothetical protein RCOM_1213430 [Ricinus communis] |
| 20 |
Hb_003025_100 |
0.104028788 |
- |
- |
PREDICTED: serine/threonine-protein phosphatase 5 isoform X2 [Jatropha curcas] |