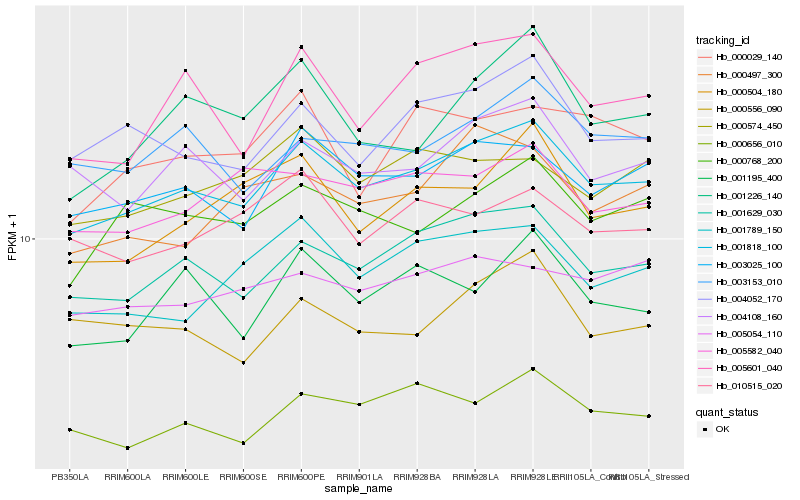
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001818_100 |
0.0 |
- |
- |
PREDICTED: DEAD-box ATP-dependent RNA helicase 11-like [Jatropha curcas] |
| 2 |
Hb_001629_030 |
0.0461929101 |
- |
- |
PAP-specific phosphatase HAL2-like family protein [Populus trichocarpa] |
| 3 |
Hb_003025_100 |
0.0576774144 |
- |
- |
PREDICTED: serine/threonine-protein phosphatase 5 isoform X2 [Jatropha curcas] |
| 4 |
Hb_004052_170 |
0.0665701117 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein PNM1, mitochondrial [Jatropha curcas] |
| 5 |
Hb_004108_160 |
0.0690550585 |
- |
- |
PREDICTED: myosin heavy chain, non-muscle-like [Jatropha curcas] |
| 6 |
Hb_005601_040 |
0.0712849007 |
- |
- |
Histone deacetylase 1 isoform 1 [Theobroma cacao] |
| 7 |
Hb_000768_200 |
0.0728110325 |
- |
- |
hypothetical protein POPTR_0013s06910g [Populus trichocarpa] |
| 8 |
Hb_001789_150 |
0.0735084357 |
- |
- |
PREDICTED: mitogen-activated protein kinase kinase kinase YODA-like isoform X1 [Jatropha curcas] |
| 9 |
Hb_003153_010 |
0.0737284821 |
- |
- |
PREDICTED: diacylglycerol kinase 3-like [Jatropha curcas] |
| 10 |
Hb_000656_010 |
0.0769615696 |
- |
- |
PREDICTED: uncharacterized protein LOC105631354 isoform X1 [Jatropha curcas] |
| 11 |
Hb_005582_040 |
0.0771362889 |
- |
- |
PREDICTED: regulator of nonsense transcripts 1 homolog [Jatropha curcas] |
| 12 |
Hb_000504_180 |
0.0781581599 |
- |
- |
PREDICTED: heterogeneous nuclear ribonucleoprotein 1 [Jatropha curcas] |
| 13 |
Hb_000574_450 |
0.0794974657 |
- |
- |
PREDICTED: signal recognition particle 54 kDa protein 2 [Jatropha curcas] |
| 14 |
Hb_001195_400 |
0.0799000375 |
- |
- |
glucose-6-phosphate isomerase, putative [Ricinus communis] |
| 15 |
Hb_005054_110 |
0.0801968306 |
- |
- |
PREDICTED: adenylosuccinate lyase-like [Jatropha curcas] |
| 16 |
Hb_000497_300 |
0.081288954 |
- |
- |
peroxisomal membrane protein [Hevea brasiliensis] |
| 17 |
Hb_000556_090 |
0.0814570624 |
- |
- |
PREDICTED: uncharacterized protein LOC105643091 isoform X1 [Jatropha curcas] |
| 18 |
Hb_010515_020 |
0.0819159185 |
- |
- |
PREDICTED: protein transport protein SEC23-like [Jatropha curcas] |
| 19 |
Hb_001226_140 |
0.0826313208 |
- |
- |
PREDICTED: protein phosphatase 2C 77-like [Jatropha curcas] |
| 20 |
Hb_000029_140 |
0.0830908676 |
- |
- |
PREDICTED: putative GDP-L-fucose synthase 2 [Populus euphratica] |