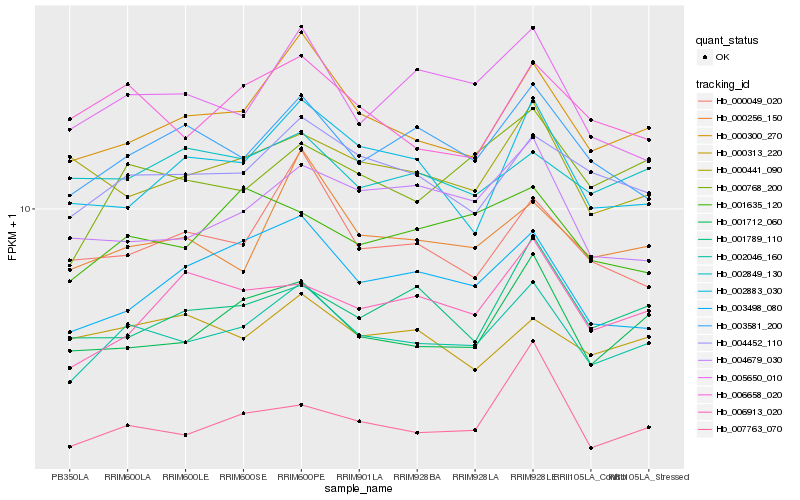
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002046_160 |
0.0 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 2 |
Hb_000300_270 |
0.0546163061 |
- |
- |
cationic amino acid transporter, putative [Ricinus communis] |
| 3 |
Hb_004452_110 |
0.0587318571 |
- |
- |
PREDICTED: uncharacterized protein LOC105639575 isoform X1 [Jatropha curcas] |
| 4 |
Hb_001712_060 |
0.0646595436 |
- |
- |
PREDICTED: proline-rich receptor-like protein kinase PERK9 [Jatropha curcas] |
| 5 |
Hb_000768_200 |
0.0680319926 |
- |
- |
hypothetical protein POPTR_0013s06910g [Populus trichocarpa] |
| 6 |
Hb_003581_200 |
0.068513656 |
- |
- |
PREDICTED: uncharacterized protein LOC105640179 [Jatropha curcas] |
| 7 |
Hb_006658_020 |
0.0717741434 |
- |
- |
PREDICTED: uncharacterized protein LOC105633529 [Jatropha curcas] |
| 8 |
Hb_000256_150 |
0.0719244329 |
- |
- |
PREDICTED: trafficking protein particle complex II-specific subunit 120 homolog [Jatropha curcas] |
| 9 |
Hb_003498_080 |
0.0753266343 |
- |
- |
PREDICTED: decapping 5-like protein isoform X2 [Jatropha curcas] |
| 10 |
Hb_005650_010 |
0.0767898319 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 11 |
Hb_001635_120 |
0.0783362976 |
- |
- |
PREDICTED: uncharacterized protein At1g51745-like [Jatropha curcas] |
| 12 |
Hb_000049_020 |
0.0791885923 |
- |
- |
Ras-GTPase-activating protein-binding protein, putative [Ricinus communis] |
| 13 |
Hb_001789_110 |
0.080134713 |
- |
- |
ubiquitin-protein ligase, putative [Ricinus communis] |
| 14 |
Hb_002883_030 |
0.080381913 |
- |
- |
PREDICTED: uncharacterized protein LOC105638222 [Jatropha curcas] |
| 15 |
Hb_006913_020 |
0.0809164607 |
- |
- |
PREDICTED: uncharacterized protein LOC105649145 isoform X1 [Jatropha curcas] |
| 16 |
Hb_000441_090 |
0.0817527666 |
- |
- |
PREDICTED: serine/threonine-protein phosphatase 6 regulatory subunit 2 [Jatropha curcas] |
| 17 |
Hb_007763_070 |
0.0818964504 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g71460, chloroplastic [Jatropha curcas] |
| 18 |
Hb_004679_030 |
0.0820924989 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: F-box protein SKIP23-like [Jatropha curcas] |
| 19 |
Hb_002849_130 |
0.0829588593 |
- |
- |
PREDICTED: RAB6A-GEF complex partner protein 1-like [Jatropha curcas] |
| 20 |
Hb_000313_220 |
0.0832145641 |
- |
- |
PREDICTED: sulfhydryl oxidase 2 isoform X1 [Jatropha curcas] |