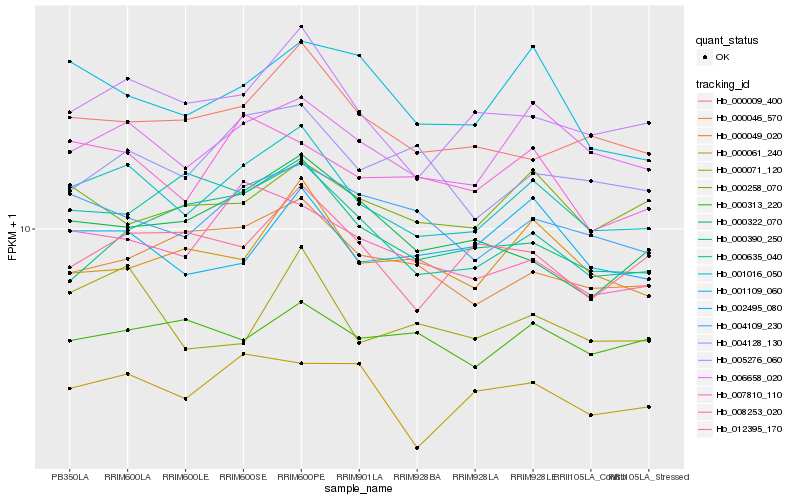
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001016_050 |
0.0 |
- |
- |
drought-inducible protein [Manihot esculenta] |
| 2 |
Hb_006658_020 |
0.0611039243 |
- |
- |
PREDICTED: uncharacterized protein LOC105633529 [Jatropha curcas] |
| 3 |
Hb_005276_060 |
0.0652140396 |
- |
- |
PREDICTED: cyclin-P3-1 isoform X2 [Jatropha curcas] |
| 4 |
Hb_000009_400 |
0.0799194428 |
- |
- |
PREDICTED: BTB/POZ domain-containing protein At3g50780 [Jatropha curcas] |
| 5 |
Hb_004109_230 |
0.0803383409 |
- |
- |
heat shock protein binding protein, putative [Ricinus communis] |
| 6 |
Hb_008253_020 |
0.0837729125 |
- |
- |
PREDICTED: serine/threonine protein phosphatase 2A 57 kDa regulatory subunit B' theta isoform-like [Jatropha curcas] |
| 7 |
Hb_007810_110 |
0.0859274875 |
- |
- |
PREDICTED: DNA/RNA-binding protein KIN17 [Jatropha curcas] |
| 8 |
Hb_000046_570 |
0.0877787793 |
- |
- |
hypothetical protein JCGZ_01998 [Jatropha curcas] |
| 9 |
Hb_004128_130 |
0.0891403675 |
- |
- |
PREDICTED: protein MODIFIER OF SNC1 11 isoform X1 [Jatropha curcas] |
| 10 |
Hb_002495_080 |
0.0894916646 |
- |
- |
PREDICTED: polygalacturonase ADPG2 [Jatropha curcas] |
| 11 |
Hb_012395_170 |
0.0905874303 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_000049_020 |
0.0936673355 |
- |
- |
Ras-GTPase-activating protein-binding protein, putative [Ricinus communis] |
| 13 |
Hb_000322_070 |
0.0940227428 |
- |
- |
PREDICTED: glycylpeptide N-tetradecanoyltransferase 1 [Jatropha curcas] |
| 14 |
Hb_000071_120 |
0.0941528296 |
- |
- |
PREDICTED: fasciclin-like arabinogalactan protein 4 [Jatropha curcas] |
| 15 |
Hb_001109_060 |
0.0947240347 |
- |
- |
amino acid transporter, putative [Ricinus communis] |
| 16 |
Hb_000061_240 |
0.09611458 |
- |
- |
kinase, putative [Ricinus communis] |
| 17 |
Hb_000258_070 |
0.0964589474 |
- |
- |
PREDICTED: uncharacterized protein LOC105649104 [Jatropha curcas] |
| 18 |
Hb_000313_220 |
0.0978364442 |
- |
- |
PREDICTED: sulfhydryl oxidase 2 isoform X1 [Jatropha curcas] |
| 19 |
Hb_000635_040 |
0.0978656596 |
- |
- |
PREDICTED: nuclear pore complex protein NUP54 [Jatropha curcas] |
| 20 |
Hb_000390_250 |
0.0983625972 |
- |
- |
PREDICTED: inositol-tetrakisphosphate 1-kinase 1 [Jatropha curcas] |