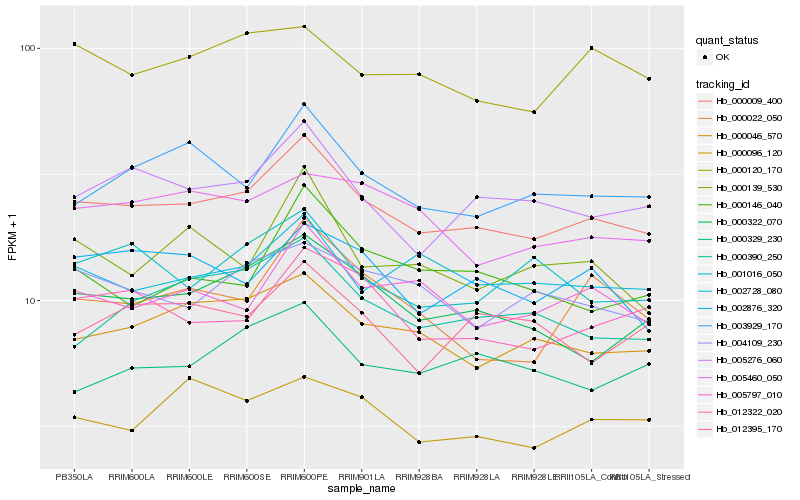
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000009_400 |
0.0 |
- |
- |
PREDICTED: BTB/POZ domain-containing protein At3g50780 [Jatropha curcas] |
| 2 |
Hb_005276_060 |
0.0641816558 |
- |
- |
PREDICTED: cyclin-P3-1 isoform X2 [Jatropha curcas] |
| 3 |
Hb_000046_570 |
0.0663798263 |
- |
- |
hypothetical protein JCGZ_01998 [Jatropha curcas] |
| 4 |
Hb_000322_070 |
0.0679434822 |
- |
- |
PREDICTED: glycylpeptide N-tetradecanoyltransferase 1 [Jatropha curcas] |
| 5 |
Hb_003929_170 |
0.0708545902 |
- |
- |
PREDICTED: novel plant SNARE 13 [Jatropha curcas] |
| 6 |
Hb_005797_010 |
0.0712737771 |
- |
- |
PREDICTED: lysophospholipid acyltransferase 1-like [Jatropha curcas] |
| 7 |
Hb_012322_020 |
0.0721484229 |
- |
- |
- |
| 8 |
Hb_004109_230 |
0.0769708993 |
- |
- |
heat shock protein binding protein, putative [Ricinus communis] |
| 9 |
Hb_002728_080 |
0.077158245 |
- |
- |
WD-repeat protein, putative [Ricinus communis] |
| 10 |
Hb_002876_320 |
0.0771609188 |
- |
- |
PREDICTED: uncharacterized protein LOC105632052 [Jatropha curcas] |
| 11 |
Hb_000120_170 |
0.079813198 |
- |
- |
selenoprotein [Populus trichocarpa] |
| 12 |
Hb_001016_050 |
0.0799194428 |
- |
- |
drought-inducible protein [Manihot esculenta] |
| 13 |
Hb_005460_050 |
0.0808089433 |
- |
- |
COR413-PM2, putative [Ricinus communis] |
| 14 |
Hb_000390_250 |
0.081070236 |
- |
- |
PREDICTED: inositol-tetrakisphosphate 1-kinase 1 [Jatropha curcas] |
| 15 |
Hb_000139_530 |
0.0836247002 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_000329_230 |
0.0837807847 |
- |
- |
PREDICTED: uncharacterized protein LOC105643152 [Jatropha curcas] |
| 17 |
Hb_000096_120 |
0.0841158054 |
- |
- |
PREDICTED: uncharacterized protein LOC105639993 [Jatropha curcas] |
| 18 |
Hb_000022_050 |
0.0844698494 |
- |
- |
PREDICTED: retinol dehydrogenase 11-like isoform X2 [Glycine max] |
| 19 |
Hb_000146_040 |
0.0849115068 |
- |
- |
PREDICTED: reticulon-like protein B11 [Jatropha curcas] |
| 20 |
Hb_012395_170 |
0.0853144441 |
- |
- |
conserved hypothetical protein [Ricinus communis] |