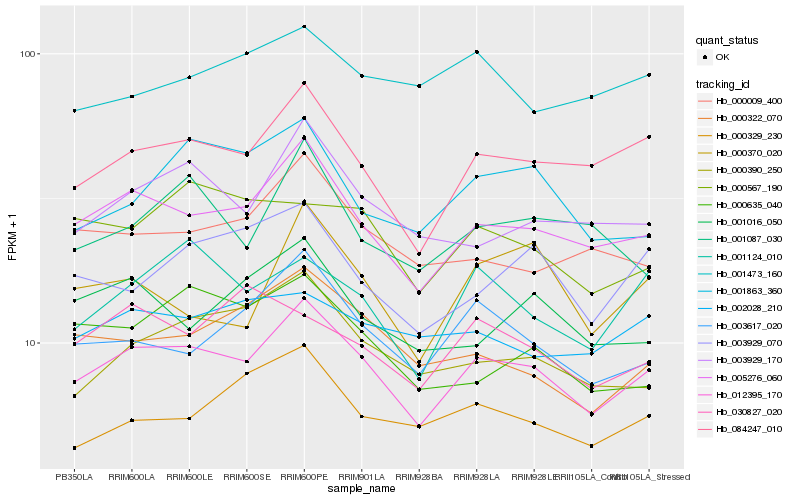
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_012395_170 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_005276_060 |
0.0455912727 |
- |
- |
PREDICTED: cyclin-P3-1 isoform X2 [Jatropha curcas] |
| 3 |
Hb_084247_010 |
0.0612745111 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_003617_020 |
0.0778112874 |
- |
- |
PREDICTED: uncharacterized protein LOC105638179 [Jatropha curcas] |
| 5 |
Hb_003929_170 |
0.0797345463 |
- |
- |
PREDICTED: novel plant SNARE 13 [Jatropha curcas] |
| 6 |
Hb_001124_010 |
0.0811992266 |
- |
- |
PREDICTED: serine/threonine-protein kinase WNK8 isoform X2 [Jatropha curcas] |
| 7 |
Hb_000390_250 |
0.0829360501 |
- |
- |
PREDICTED: inositol-tetrakisphosphate 1-kinase 1 [Jatropha curcas] |
| 8 |
Hb_000329_230 |
0.0841591434 |
- |
- |
PREDICTED: uncharacterized protein LOC105643152 [Jatropha curcas] |
| 9 |
Hb_000322_070 |
0.0851325126 |
- |
- |
PREDICTED: glycylpeptide N-tetradecanoyltransferase 1 [Jatropha curcas] |
| 10 |
Hb_000009_400 |
0.0853144441 |
- |
- |
PREDICTED: BTB/POZ domain-containing protein At3g50780 [Jatropha curcas] |
| 11 |
Hb_003929_070 |
0.0856499046 |
- |
- |
CBL-interacting serine/threonine-protein kinase 1 isoform 3 [Theobroma cacao] |
| 12 |
Hb_002028_210 |
0.0878947148 |
- |
- |
PREDICTED: uncharacterized protein LOC105638038 [Jatropha curcas] |
| 13 |
Hb_001863_360 |
0.0895518043 |
- |
- |
PREDICTED: 5'-adenylylsulfate reductase-like 4 [Jatropha curcas] |
| 14 |
Hb_001016_050 |
0.0905874303 |
- |
- |
drought-inducible protein [Manihot esculenta] |
| 15 |
Hb_000567_190 |
0.0906733024 |
- |
- |
PREDICTED: protein SCAR3 isoform X2 [Jatropha curcas] |
| 16 |
Hb_001087_030 |
0.0932032542 |
- |
- |
PREDICTED: serine/threonine-protein phosphatase PP2A catalytic subunit [Jatropha curcas] |
| 17 |
Hb_000370_020 |
0.0942995916 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_001473_160 |
0.0949789476 |
- |
- |
Vesicle-associated membrane protein, putative [Ricinus communis] |
| 19 |
Hb_000635_040 |
0.0955847089 |
- |
- |
PREDICTED: nuclear pore complex protein NUP54 [Jatropha curcas] |
| 20 |
Hb_030827_020 |
0.0961095029 |
- |
- |
PREDICTED: protein ROOT PRIMORDIUM DEFECTIVE 1 [Jatropha curcas] |