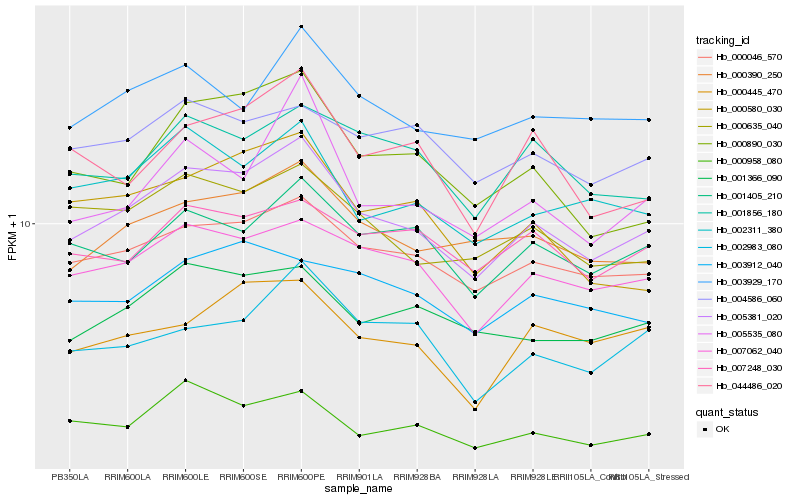
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_005381_020 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105630417 [Jatropha curcas] |
| 2 |
Hb_000046_570 |
0.060929284 |
- |
- |
hypothetical protein JCGZ_01998 [Jatropha curcas] |
| 3 |
Hb_007062_040 |
0.0685000456 |
- |
- |
ADP-ribosylation factor GTPase-activating protein AGD2 [Morus notabilis] |
| 4 |
Hb_000390_250 |
0.0718993833 |
- |
- |
PREDICTED: inositol-tetrakisphosphate 1-kinase 1 [Jatropha curcas] |
| 5 |
Hb_000635_040 |
0.0784102092 |
- |
- |
PREDICTED: nuclear pore complex protein NUP54 [Jatropha curcas] |
| 6 |
Hb_001366_090 |
0.0784403227 |
transcription factor |
TF Family: NAC |
hypothetical protein RCOM_1341380 [Ricinus communis] |
| 7 |
Hb_002311_380 |
0.079276683 |
- |
- |
PREDICTED: induced during hyphae development protein 1 [Jatropha curcas] |
| 8 |
Hb_001405_210 |
0.0818781287 |
- |
- |
hypothetical protein JCGZ_22540 [Jatropha curcas] |
| 9 |
Hb_000890_030 |
0.0821225057 |
- |
- |
plastid CUL1 [Hevea brasiliensis] |
| 10 |
Hb_003929_170 |
0.0823145167 |
- |
- |
PREDICTED: novel plant SNARE 13 [Jatropha curcas] |
| 11 |
Hb_001856_180 |
0.0883818613 |
- |
- |
PREDICTED: eukaryotic translation initiation factor 3 subunit M [Jatropha curcas] |
| 12 |
Hb_005535_080 |
0.0892690584 |
- |
- |
hypothetical protein CICLE_v10012766mg [Citrus clementina] |
| 13 |
Hb_044486_020 |
0.0897982801 |
- |
- |
CASTOR protein [Glycine max] |
| 14 |
Hb_000958_080 |
0.0902408492 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_000445_470 |
0.0913807721 |
- |
- |
PREDICTED: RING finger and transmembrane domain-containing protein 1-like [Jatropha curcas] |
| 16 |
Hb_007248_030 |
0.0916780118 |
- |
- |
PREDICTED: uncharacterized protein LOC105633558 [Jatropha curcas] |
| 17 |
Hb_004586_060 |
0.0917023355 |
- |
- |
PREDICTED: AP-4 complex subunit mu [Jatropha curcas] |
| 18 |
Hb_002983_080 |
0.0919360419 |
- |
- |
acetylornithine aminotransferase, partial [Prunus persica] |
| 19 |
Hb_003912_040 |
0.0926875072 |
- |
- |
hypothetical protein CISIN_1g005153mg [Citrus sinensis] |
| 20 |
Hb_000580_030 |
0.093580706 |
- |
- |
PREDICTED: probable protein S-acyltransferase 14 [Jatropha curcas] |