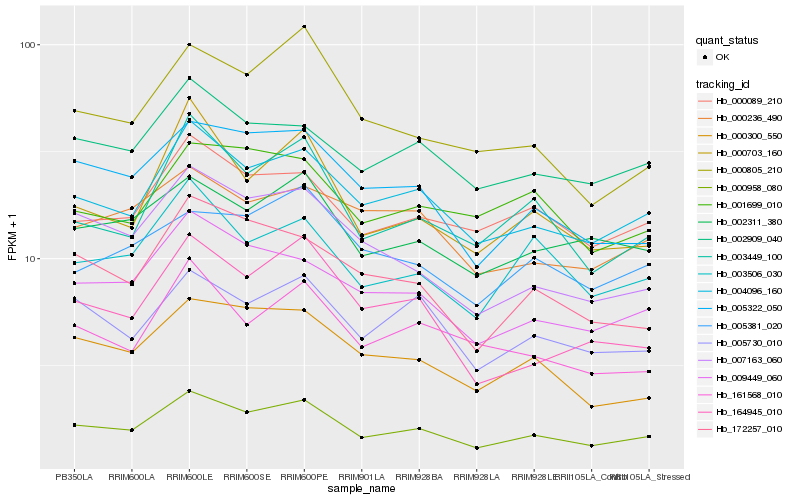
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000958_080 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_004096_160 |
0.0459936318 |
- |
- |
PREDICTED: T-complex protein 1 subunit zeta 1 [Jatropha curcas] |
| 3 |
Hb_009449_060 |
0.0681605594 |
- |
- |
PREDICTED: ubiquitin-like-specific protease 1D isoform X2 [Jatropha curcas] |
| 4 |
Hb_005322_050 |
0.0720672825 |
- |
- |
hypothetical protein POPTR_0009s02400g [Populus trichocarpa] |
| 5 |
Hb_000089_210 |
0.0735306395 |
- |
- |
unknown [Medicago truncatula] |
| 6 |
Hb_002909_040 |
0.0737146047 |
desease resistance |
Gene Name: AAA |
PREDICTED: 26S protease regulatory subunit 6B homolog [Jatropha curcas] |
| 7 |
Hb_005730_010 |
0.0758809519 |
- |
- |
PREDICTED: ubiquitin carboxyl-terminal hydrolase 7 isoform X1 [Populus euphratica] |
| 8 |
Hb_002311_380 |
0.076220328 |
- |
- |
PREDICTED: induced during hyphae development protein 1 [Jatropha curcas] |
| 9 |
Hb_003449_100 |
0.0786672961 |
- |
- |
PREDICTED: protein YIPF6 homolog [Jatropha curcas] |
| 10 |
Hb_000300_550 |
0.0799002248 |
- |
- |
PREDICTED: uncharacterized protein At1g04910-like [Jatropha curcas] |
| 11 |
Hb_003506_030 |
0.0811357675 |
- |
- |
PREDICTED: ATP-dependent Clp protease ATP-binding subunit clpX-like, mitochondrial isoform X2 [Jatropha curcas] |
| 12 |
Hb_007163_060 |
0.0828372462 |
- |
- |
PREDICTED: protein RMD5 homolog A [Jatropha curcas] |
| 13 |
Hb_161568_010 |
0.0843956135 |
- |
- |
hydrolase, hydrolyzing O-glycosyl compounds, putative [Ricinus communis] |
| 14 |
Hb_172257_010 |
0.0855233721 |
transcription factor |
TF Family: HMG |
high mobility group family protein [Populus trichocarpa] |
| 15 |
Hb_000703_160 |
0.087481541 |
- |
- |
PREDICTED: serine/threonine-protein kinase EDR1 isoform X2 [Jatropha curcas] |
| 16 |
Hb_000805_210 |
0.0881033987 |
- |
- |
PREDICTED: oxysterol-binding protein-related protein 3C-like [Jatropha curcas] |
| 17 |
Hb_000236_490 |
0.089586483 |
- |
- |
20 kD nuclear cap binding protein, putative [Ricinus communis] |
| 18 |
Hb_164945_010 |
0.0899290092 |
- |
- |
PREDICTED: DNA primase small subunit [Jatropha curcas] |
| 19 |
Hb_005381_020 |
0.0902408492 |
- |
- |
PREDICTED: uncharacterized protein LOC105630417 [Jatropha curcas] |
| 20 |
Hb_001699_010 |
0.0907443906 |
- |
- |
drought-inducible protein [Manihot esculenta] |