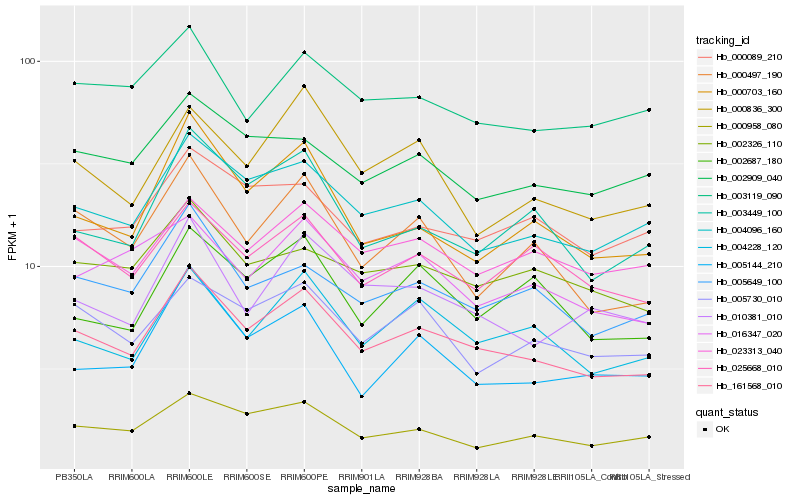
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_161568_010 |
0.0 |
- |
- |
hydrolase, hydrolyzing O-glycosyl compounds, putative [Ricinus communis] |
| 2 |
Hb_004096_160 |
0.0710802485 |
- |
- |
PREDICTED: T-complex protein 1 subunit zeta 1 [Jatropha curcas] |
| 3 |
Hb_004228_120 |
0.0817244633 |
- |
- |
hypothetical protein POPTR_0013s02080g [Populus trichocarpa] |
| 4 |
Hb_005649_100 |
0.0829108414 |
- |
- |
Ras-GTPase-activating protein-binding protein, putative [Ricinus communis] |
| 5 |
Hb_000958_080 |
0.0843956135 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_000703_160 |
0.0873834551 |
- |
- |
PREDICTED: serine/threonine-protein kinase EDR1 isoform X2 [Jatropha curcas] |
| 7 |
Hb_000497_190 |
0.0892875745 |
- |
- |
sterol isomerase, putative [Ricinus communis] |
| 8 |
Hb_010381_010 |
0.0896327021 |
- |
- |
WD-repeat protein, putative [Ricinus communis] |
| 9 |
Hb_023313_040 |
0.0911630898 |
- |
- |
PREDICTED: uncharacterized protein LOC105640827 isoform X2 [Jatropha curcas] |
| 10 |
Hb_025668_010 |
0.0917675807 |
- |
- |
unnamed protein product [Coffea canephora] |
| 11 |
Hb_003449_100 |
0.0921284228 |
- |
- |
PREDICTED: protein YIPF6 homolog [Jatropha curcas] |
| 12 |
Hb_002326_110 |
0.0921698561 |
- |
- |
PREDICTED: uncharacterized protein LOC103320920 [Prunus mume] |
| 13 |
Hb_005730_010 |
0.0925446418 |
- |
- |
PREDICTED: ubiquitin carboxyl-terminal hydrolase 7 isoform X1 [Populus euphratica] |
| 14 |
Hb_003119_090 |
0.0958270371 |
- |
- |
PREDICTED: 14-3-3 protein 6 [Jatropha curcas] |
| 15 |
Hb_016347_020 |
0.0975112193 |
- |
- |
PREDICTED: D-amino-acid transaminase, chloroplastic-like isoform X1 [Populus euphratica] |
| 16 |
Hb_002909_040 |
0.0981246557 |
desease resistance |
Gene Name: AAA |
PREDICTED: 26S protease regulatory subunit 6B homolog [Jatropha curcas] |
| 17 |
Hb_000089_210 |
0.0982633926 |
- |
- |
unknown [Medicago truncatula] |
| 18 |
Hb_000836_300 |
0.0985073381 |
- |
- |
caax prenyl protease ste24, putative [Ricinus communis] |
| 19 |
Hb_005144_210 |
0.0996274496 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_002687_180 |
0.1000551871 |
- |
- |
PREDICTED: phytochrome-associated serine/threonine-protein phosphatase [Jatropha curcas] |