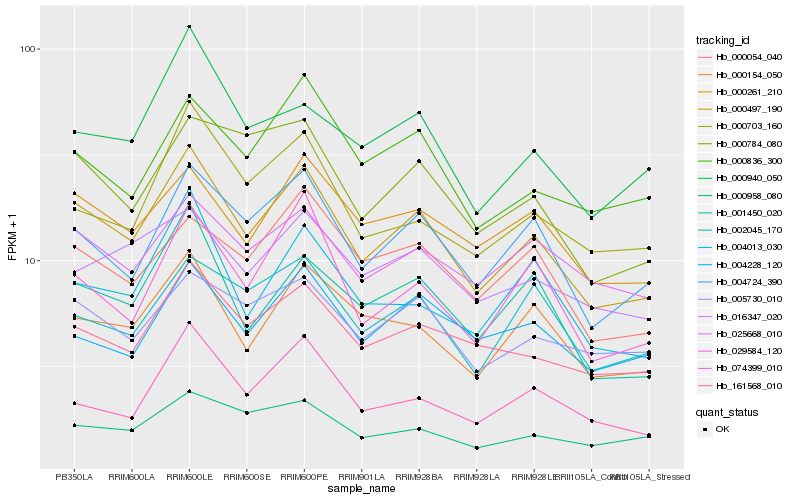
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000497_190 |
0.0 |
- |
- |
sterol isomerase, putative [Ricinus communis] |
| 2 |
Hb_004724_390 |
0.0800017968 |
- |
- |
PREDICTED: nicalin-1-like [Populus euphratica] |
| 3 |
Hb_161568_010 |
0.0892875745 |
- |
- |
hydrolase, hydrolyzing O-glycosyl compounds, putative [Ricinus communis] |
| 4 |
Hb_000261_210 |
0.0924929964 |
- |
- |
PREDICTED: uncharacterized protein At1g04910 [Jatropha curcas] |
| 5 |
Hb_000154_050 |
0.0928356314 |
- |
- |
CMP-sialic acid transporter, putative [Ricinus communis] |
| 6 |
Hb_001450_020 |
0.0933177325 |
- |
- |
PREDICTED: pyruvate kinase isozyme A, chloroplastic-like [Jatropha curcas] |
| 7 |
Hb_005730_010 |
0.0983825119 |
- |
- |
PREDICTED: ubiquitin carboxyl-terminal hydrolase 7 isoform X1 [Populus euphratica] |
| 8 |
Hb_025668_010 |
0.0995333103 |
- |
- |
unnamed protein product [Coffea canephora] |
| 9 |
Hb_000784_080 |
0.0995935059 |
- |
- |
protein disulfide isomerase, putative [Ricinus communis] |
| 10 |
Hb_029584_120 |
0.1004075226 |
- |
- |
PREDICTED: K(+) efflux antiporter 4 isoform X1 [Jatropha curcas] |
| 11 |
Hb_000836_300 |
0.1017641752 |
- |
- |
caax prenyl protease ste24, putative [Ricinus communis] |
| 12 |
Hb_000703_160 |
0.1020939482 |
- |
- |
PREDICTED: serine/threonine-protein kinase EDR1 isoform X2 [Jatropha curcas] |
| 13 |
Hb_000054_040 |
0.10325845 |
- |
- |
PREDICTED: mannosyl-oligosaccharide 1,2-alpha-mannosidase MNS1 [Jatropha curcas] |
| 14 |
Hb_000940_050 |
0.1041433345 |
- |
- |
hypothetical protein JCGZ_19588 [Jatropha curcas] |
| 15 |
Hb_000958_080 |
0.1041783438 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_004013_030 |
0.1044930158 |
- |
- |
Phosphatidylinositol-3,4,5-trisphosphate 3-phosphatase and dual-specificity protein phosphatase PTEN, putative [Ricinus communis] |
| 17 |
Hb_002045_170 |
0.1047456372 |
- |
- |
interferon-induced guanylate-binding protein, putative [Ricinus communis] |
| 18 |
Hb_074399_010 |
0.1053070142 |
- |
- |
PREDICTED: phragmoplast orienting kinesin-1 isoform X1 [Populus euphratica] |
| 19 |
Hb_004228_120 |
0.1063947667 |
- |
- |
hypothetical protein POPTR_0013s02080g [Populus trichocarpa] |
| 20 |
Hb_016347_020 |
0.1070561245 |
- |
- |
PREDICTED: D-amino-acid transaminase, chloroplastic-like isoform X1 [Populus euphratica] |