| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
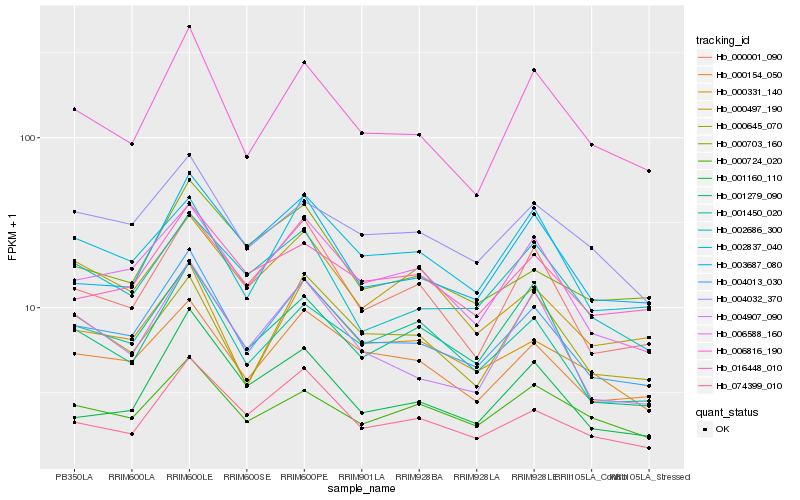
Hb_004013_030 |
0.0 |
- |
- |
Phosphatidylinositol-3,4,5-trisphosphate 3-phosphatase and dual-specificity protein phosphatase PTEN, putative [Ricinus communis] |
| 2 |
Hb_006588_160 |
0.0732176868 |
- |
- |
PREDICTED: acetyl-coenzyme A carboxylase carboxyl transferase subunit alpha, chloroplastic isoform X2 [Jatropha curcas] |
| 3 |
Hb_000154_050 |
0.0739043191 |
- |
- |
CMP-sialic acid transporter, putative [Ricinus communis] |
| 4 |
Hb_006816_190 |
0.0767801856 |
- |
- |
calreticulin family protein [Populus trichocarpa] |
| 5 |
Hb_001450_020 |
0.0791382473 |
- |
- |
PREDICTED: pyruvate kinase isozyme A, chloroplastic-like [Jatropha curcas] |
| 6 |
Hb_000331_140 |
0.0815151559 |
- |
- |
PREDICTED: cycloeucalenol cycloisomerase [Jatropha curcas] |
| 7 |
Hb_074399_010 |
0.0858862193 |
- |
- |
PREDICTED: phragmoplast orienting kinesin-1 isoform X1 [Populus euphratica] |
| 8 |
Hb_000001_090 |
0.0939891391 |
- |
- |
PREDICTED: uncharacterized protein At3g58460 [Populus euphratica] |
| 9 |
Hb_004032_370 |
0.0941785378 |
- |
- |
pyrophosphate-dependent phosphofructokinase [Hevea brasiliensis] |
| 10 |
Hb_004907_090 |
0.0981728418 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 11 |
Hb_003687_080 |
0.1007827056 |
- |
- |
PREDICTED: casein kinase I-like [Jatropha curcas] |
| 12 |
Hb_016448_010 |
0.1012263531 |
- |
- |
Protein MYG1, putative [Ricinus communis] |
| 13 |
Hb_002837_040 |
0.1033617835 |
- |
- |
Transmembrane emp24 domain-containing protein 10 precursor, putative [Ricinus communis] |
| 14 |
Hb_002686_300 |
0.1041975066 |
- |
- |
adenosine diphosphatase, putative [Ricinus communis] |
| 15 |
Hb_000497_190 |
0.1044930158 |
- |
- |
sterol isomerase, putative [Ricinus communis] |
| 16 |
Hb_001279_090 |
0.1068476728 |
- |
- |
PREDICTED: uncharacterized protein LOC105633322 [Jatropha curcas] |
| 17 |
Hb_000724_020 |
0.1068700907 |
transcription factor |
TF Family: PHD |
PREDICTED: probable Histone-lysine N-methyltransferase ATXR5 [Jatropha curcas] |
| 18 |
Hb_000703_160 |
0.1075931621 |
- |
- |
PREDICTED: serine/threonine-protein kinase EDR1 isoform X2 [Jatropha curcas] |
| 19 |
Hb_001160_110 |
0.1085201208 |
- |
- |
PREDICTED: kinesin-related protein 13 [Jatropha curcas] |
| 20 |
Hb_000645_070 |
0.1085672405 |
- |
- |
aldehyde dehydrogenase, putative [Ricinus communis] |