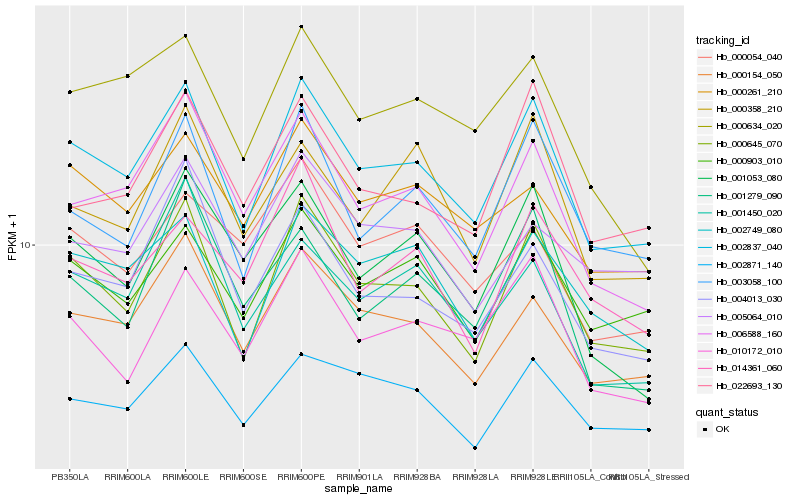
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002837_040 |
0.0 |
- |
- |
Transmembrane emp24 domain-containing protein 10 precursor, putative [Ricinus communis] |
| 2 |
Hb_000645_070 |
0.0480476215 |
- |
- |
aldehyde dehydrogenase, putative [Ricinus communis] |
| 3 |
Hb_000903_010 |
0.0767793366 |
- |
- |
hypothetical protein JCGZ_23825 [Jatropha curcas] |
| 4 |
Hb_000154_050 |
0.0768489688 |
- |
- |
CMP-sialic acid transporter, putative [Ricinus communis] |
| 5 |
Hb_002749_080 |
0.081639155 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_003058_100 |
0.0829163096 |
- |
- |
PREDICTED: hydroxymethylglutaryl-CoA lyase, mitochondrial [Jatropha curcas] |
| 7 |
Hb_006588_160 |
0.0846131421 |
- |
- |
PREDICTED: acetyl-coenzyme A carboxylase carboxyl transferase subunit alpha, chloroplastic isoform X2 [Jatropha curcas] |
| 8 |
Hb_002871_140 |
0.0883382717 |
- |
- |
hypothetical protein JCGZ_06847 [Jatropha curcas] |
| 9 |
Hb_010172_010 |
0.0884543544 |
- |
- |
PREDICTED: probable beta-1,3-galactosyltransferase 20 [Jatropha curcas] |
| 10 |
Hb_000634_020 |
0.0930422834 |
- |
- |
hypothetical protein JCGZ_11888 [Jatropha curcas] |
| 11 |
Hb_000261_210 |
0.0935390996 |
- |
- |
PREDICTED: uncharacterized protein At1g04910 [Jatropha curcas] |
| 12 |
Hb_022693_130 |
0.0976788605 |
- |
- |
BRASSINOSTEROID INSENSITIVE 1-associated receptor kinase 1 precursor, putative [Ricinus communis] |
| 13 |
Hb_001053_080 |
0.1018167173 |
- |
- |
OsCesA3 protein [Morus notabilis] |
| 14 |
Hb_004013_030 |
0.1033617835 |
- |
- |
Phosphatidylinositol-3,4,5-trisphosphate 3-phosphatase and dual-specificity protein phosphatase PTEN, putative [Ricinus communis] |
| 15 |
Hb_014361_060 |
0.105188621 |
- |
- |
PREDICTED: probable prolyl 4-hydroxylase 9 [Jatropha curcas] |
| 16 |
Hb_000358_210 |
0.1071939775 |
- |
- |
oligosaccharyl transferase, putative [Ricinus communis] |
| 17 |
Hb_005064_010 |
0.1110956241 |
- |
- |
PREDICTED: kelch repeat-containing protein At3g27220-like [Jatropha curcas] |
| 18 |
Hb_000054_040 |
0.1115400435 |
- |
- |
PREDICTED: mannosyl-oligosaccharide 1,2-alpha-mannosidase MNS1 [Jatropha curcas] |
| 19 |
Hb_001279_090 |
0.1120676829 |
- |
- |
PREDICTED: uncharacterized protein LOC105633322 [Jatropha curcas] |
| 20 |
Hb_001450_020 |
0.1122265203 |
- |
- |
PREDICTED: pyruvate kinase isozyme A, chloroplastic-like [Jatropha curcas] |