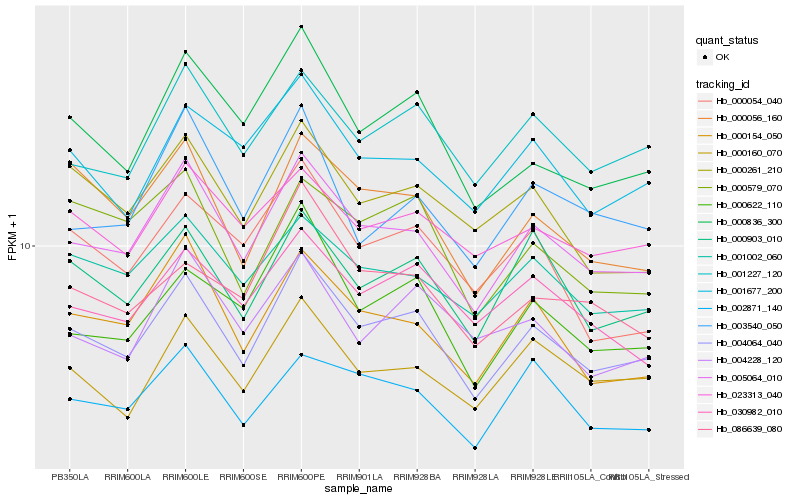
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004064_040 |
0.0 |
- |
- |
PREDICTED: Down syndrome critical region protein 3 homolog isoform X3 [Jatropha curcas] |
| 2 |
Hb_005064_010 |
0.0458131604 |
- |
- |
PREDICTED: kelch repeat-containing protein At3g27220-like [Jatropha curcas] |
| 3 |
Hb_000836_300 |
0.0766553374 |
- |
- |
caax prenyl protease ste24, putative [Ricinus communis] |
| 4 |
Hb_000903_010 |
0.0774292469 |
- |
- |
hypothetical protein JCGZ_23825 [Jatropha curcas] |
| 5 |
Hb_000160_070 |
0.081585972 |
- |
- |
PREDICTED: uncharacterized protein LOC105648219 [Jatropha curcas] |
| 6 |
Hb_000056_160 |
0.0817426119 |
- |
- |
PREDICTED: peptidyl-prolyl cis-trans isomerase CYP23 isoform X1 [Jatropha curcas] |
| 7 |
Hb_001227_120 |
0.0825277475 |
transcription factor |
TF Family: C2H2 |
Histone deacetylase 2a, putative [Ricinus communis] |
| 8 |
Hb_000622_110 |
0.0869578782 |
- |
- |
cmp-sialic acid transporter, putative [Ricinus communis] |
| 9 |
Hb_001677_200 |
0.0870545899 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_000154_050 |
0.0872743215 |
- |
- |
CMP-sialic acid transporter, putative [Ricinus communis] |
| 11 |
Hb_000261_210 |
0.0880827092 |
- |
- |
PREDICTED: uncharacterized protein At1g04910 [Jatropha curcas] |
| 12 |
Hb_001002_060 |
0.0893730997 |
- |
- |
PREDICTED: putative GPI-anchor transamidase [Jatropha curcas] |
| 13 |
Hb_030982_010 |
0.0933822457 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_023313_040 |
0.0946905933 |
- |
- |
PREDICTED: uncharacterized protein LOC105640827 isoform X2 [Jatropha curcas] |
| 15 |
Hb_004228_120 |
0.0966194201 |
- |
- |
hypothetical protein POPTR_0013s02080g [Populus trichocarpa] |
| 16 |
Hb_086639_080 |
0.0974818551 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_002871_140 |
0.0975809742 |
- |
- |
hypothetical protein JCGZ_06847 [Jatropha curcas] |
| 18 |
Hb_000054_040 |
0.0979187006 |
- |
- |
PREDICTED: mannosyl-oligosaccharide 1,2-alpha-mannosidase MNS1 [Jatropha curcas] |
| 19 |
Hb_000579_070 |
0.0984748783 |
- |
- |
hypothetical protein CISIN_1g0119542mg, partial [Citrus sinensis] |
| 20 |
Hb_003540_050 |
0.0986750476 |
- |
- |
PREDICTED: plant intracellular Ras-group-related LRR protein 6 isoform X4 [Jatropha curcas] |