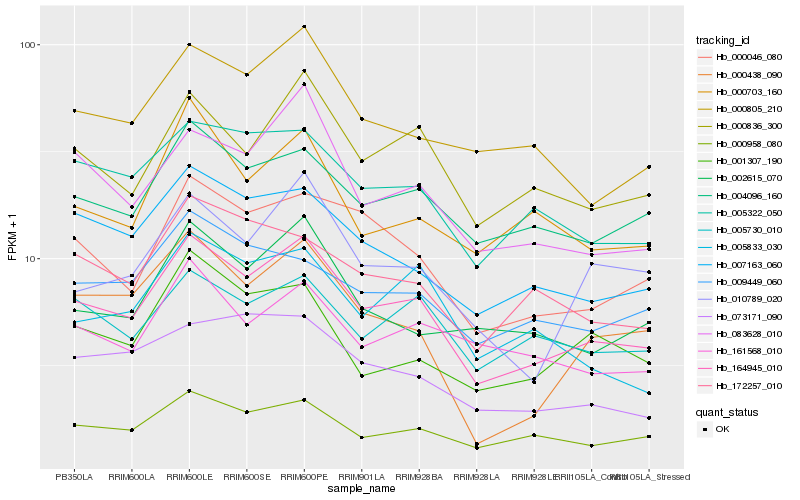
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_164945_010 |
0.0 |
- |
- |
PREDICTED: DNA primase small subunit [Jatropha curcas] |
| 2 |
Hb_000836_300 |
0.0884709477 |
- |
- |
caax prenyl protease ste24, putative [Ricinus communis] |
| 3 |
Hb_000958_080 |
0.0899290092 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_000046_080 |
0.0927563506 |
- |
- |
PREDICTED: serine/arginine-rich SC35-like splicing factor SCL28 [Jatropha curcas] |
| 5 |
Hb_007163_060 |
0.0928295325 |
- |
- |
PREDICTED: protein RMD5 homolog A [Jatropha curcas] |
| 6 |
Hb_005322_050 |
0.0951071642 |
- |
- |
hypothetical protein POPTR_0009s02400g [Populus trichocarpa] |
| 7 |
Hb_083628_010 |
0.095562028 |
- |
- |
PREDICTED: probable serine protease EDA2 [Jatropha curcas] |
| 8 |
Hb_004096_160 |
0.0958435647 |
- |
- |
PREDICTED: T-complex protein 1 subunit zeta 1 [Jatropha curcas] |
| 9 |
Hb_000805_210 |
0.1012440973 |
- |
- |
PREDICTED: oxysterol-binding protein-related protein 3C-like [Jatropha curcas] |
| 10 |
Hb_005730_010 |
0.1084678564 |
- |
- |
PREDICTED: ubiquitin carboxyl-terminal hydrolase 7 isoform X1 [Populus euphratica] |
| 11 |
Hb_002615_070 |
0.1089086262 |
- |
- |
PREDICTED: uncharacterized protein LOC105628992 isoform X1 [Jatropha curcas] |
| 12 |
Hb_073171_090 |
0.1110988984 |
- |
- |
PREDICTED: uncharacterized protein LOC105631539 [Jatropha curcas] |
| 13 |
Hb_009449_060 |
0.1115251777 |
- |
- |
PREDICTED: ubiquitin-like-specific protease 1D isoform X2 [Jatropha curcas] |
| 14 |
Hb_172257_010 |
0.1115624883 |
transcription factor |
TF Family: HMG |
high mobility group family protein [Populus trichocarpa] |
| 15 |
Hb_010789_020 |
0.1122789512 |
- |
- |
ubiquitin-conjugating enzyme e2S, putative [Ricinus communis] |
| 16 |
Hb_161568_010 |
0.1129448978 |
- |
- |
hydrolase, hydrolyzing O-glycosyl compounds, putative [Ricinus communis] |
| 17 |
Hb_000703_160 |
0.1144520461 |
- |
- |
PREDICTED: serine/threonine-protein kinase EDR1 isoform X2 [Jatropha curcas] |
| 18 |
Hb_000438_090 |
0.1167804494 |
- |
- |
PREDICTED: mini-chromosome maintenance complex-binding protein [Jatropha curcas] |
| 19 |
Hb_001307_190 |
0.1167829602 |
transcription factor |
TF Family: G2-like |
transcription factor, putative [Ricinus communis] |
| 20 |
Hb_005833_030 |
0.1169001384 |
- |
- |
PREDICTED: uncharacterized membrane protein At1g16860 [Jatropha curcas] |