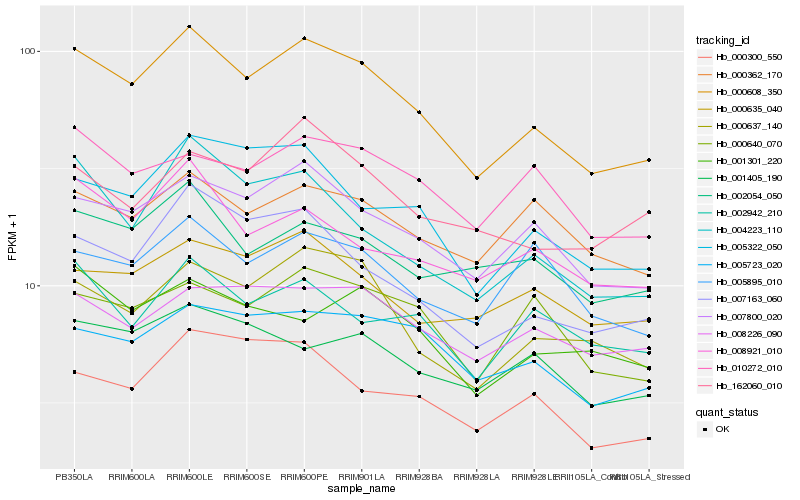
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000608_350 |
0.0 |
- |
- |
PREDICTED: ER membrane protein complex subunit 10 [Jatropha curcas] |
| 2 |
Hb_007800_020 |
0.0687401598 |
- |
- |
PREDICTED: probable arabinosyltransferase ARAD1 isoform X1 [Jatropha curcas] |
| 3 |
Hb_000637_140 |
0.0766587661 |
- |
- |
PREDICTED: actin-related protein 4-like [Jatropha curcas] |
| 4 |
Hb_007163_060 |
0.0817200212 |
- |
- |
PREDICTED: protein RMD5 homolog A [Jatropha curcas] |
| 5 |
Hb_005322_050 |
0.0855744245 |
- |
- |
hypothetical protein POPTR_0009s02400g [Populus trichocarpa] |
| 6 |
Hb_004223_110 |
0.0873353969 |
- |
- |
PREDICTED: putative HVA22-like protein g [Populus euphratica] |
| 7 |
Hb_005723_020 |
0.088231471 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_008921_010 |
0.0931171198 |
- |
- |
PREDICTED: uncharacterized protein LOC105629323 [Jatropha curcas] |
| 9 |
Hb_002054_050 |
0.0945403431 |
- |
- |
PREDICTED: uncharacterized protein LOC105641285 [Jatropha curcas] |
| 10 |
Hb_000640_070 |
0.0955633092 |
- |
- |
PREDICTED: vacuolar protein sorting-associated protein 51 homolog [Jatropha curcas] |
| 11 |
Hb_000635_040 |
0.0966808517 |
- |
- |
PREDICTED: nuclear pore complex protein NUP54 [Jatropha curcas] |
| 12 |
Hb_010272_010 |
0.0970162533 |
- |
- |
PREDICTED: endoplasmic reticulum-Golgi intermediate compartment protein 3 [Jatropha curcas] |
| 13 |
Hb_000300_550 |
0.0974270054 |
- |
- |
PREDICTED: uncharacterized protein At1g04910-like [Jatropha curcas] |
| 14 |
Hb_002942_210 |
0.0975248424 |
- |
- |
alpha-2,8-sialyltransferase 8b, putative [Ricinus communis] |
| 15 |
Hb_008226_090 |
0.0975810399 |
- |
- |
PREDICTED: reticulon-like protein B16 isoform X1 [Jatropha curcas] |
| 16 |
Hb_162060_010 |
0.0980563984 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_001405_190 |
0.0982206486 |
- |
- |
PREDICTED: NADH dehydrogenase [ubiquinone] iron-sulfur protein 1, mitochondrial [Jatropha curcas] |
| 18 |
Hb_005895_010 |
0.0988664302 |
- |
- |
PREDICTED: uncharacterized protein LOC105643387 isoform X1 [Jatropha curcas] |
| 19 |
Hb_001301_220 |
0.0995443307 |
- |
- |
PREDICTED: polyadenylate-binding protein-interacting protein 4 [Jatropha curcas] |
| 20 |
Hb_000362_170 |
0.0996335194 |
- |
- |
PREDICTED: uncharacterized protein LOC105635728 [Jatropha curcas] |