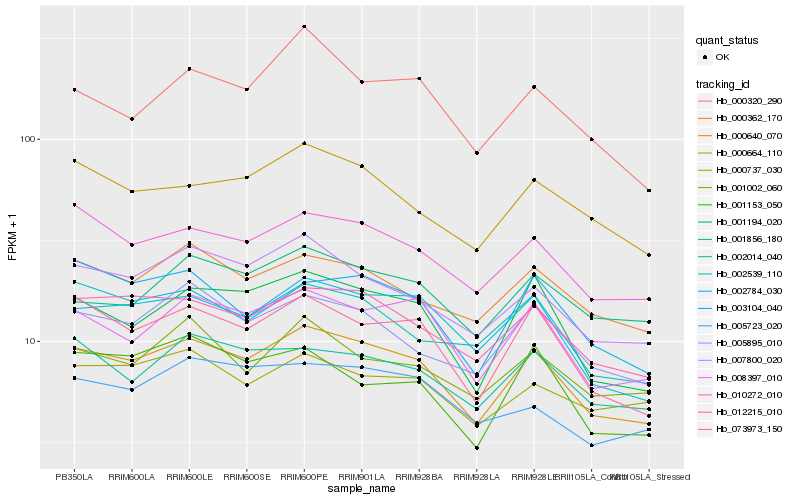
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000640_070 |
0.0 |
- |
- |
PREDICTED: vacuolar protein sorting-associated protein 51 homolog [Jatropha curcas] |
| 2 |
Hb_012215_010 |
0.0546379712 |
- |
- |
PREDICTED: YTH domain-containing family protein 1 [Populus euphratica] |
| 3 |
Hb_073973_150 |
0.0566093341 |
- |
- |
PREDICTED: aminoacylase-1 isoform X1 [Jatropha curcas] |
| 4 |
Hb_008397_010 |
0.0580129854 |
- |
- |
PREDICTED: uncharacterized protein LOC105640192 isoform X2 [Jatropha curcas] |
| 5 |
Hb_002784_030 |
0.0582312293 |
- |
- |
hypothetical protein POPTR_0001s06740g [Populus trichocarpa] |
| 6 |
Hb_010272_010 |
0.0591653206 |
- |
- |
PREDICTED: endoplasmic reticulum-Golgi intermediate compartment protein 3 [Jatropha curcas] |
| 7 |
Hb_001194_020 |
0.0612187503 |
- |
- |
microtubule associated protein xmap215, putative [Ricinus communis] |
| 8 |
Hb_007800_020 |
0.0629699582 |
- |
- |
PREDICTED: probable arabinosyltransferase ARAD1 isoform X1 [Jatropha curcas] |
| 9 |
Hb_005895_010 |
0.0682333307 |
- |
- |
PREDICTED: uncharacterized protein LOC105643387 isoform X1 [Jatropha curcas] |
| 10 |
Hb_000362_170 |
0.0702787121 |
- |
- |
PREDICTED: uncharacterized protein LOC105635728 [Jatropha curcas] |
| 11 |
Hb_000664_110 |
0.0718383029 |
- |
- |
PREDICTED: monoglyceride lipase [Jatropha curcas] |
| 12 |
Hb_002014_040 |
0.0734145127 |
- |
- |
site-1 protease, putative [Ricinus communis] |
| 13 |
Hb_003104_040 |
0.0753215945 |
- |
- |
PREDICTED: insulin-degrading enzyme isoform X1 [Jatropha curcas] |
| 14 |
Hb_002539_110 |
0.0756473029 |
- |
- |
hypothetical protein JCGZ_15697 [Jatropha curcas] |
| 15 |
Hb_000320_290 |
0.0792212683 |
- |
- |
PREDICTED: putative lactoylglutathione lyase [Jatropha curcas] |
| 16 |
Hb_005723_020 |
0.0808540232 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_001002_060 |
0.0812773316 |
- |
- |
PREDICTED: putative GPI-anchor transamidase [Jatropha curcas] |
| 18 |
Hb_001856_180 |
0.081847024 |
- |
- |
PREDICTED: eukaryotic translation initiation factor 3 subunit M [Jatropha curcas] |
| 19 |
Hb_000737_030 |
0.0820580647 |
- |
- |
PREDICTED: probable galacturonosyltransferase 3 isoform X1 [Jatropha curcas] |
| 20 |
Hb_001153_050 |
0.0843371952 |
- |
- |
PREDICTED: protein FIZZY-RELATED 2 [Jatropha curcas] |