| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
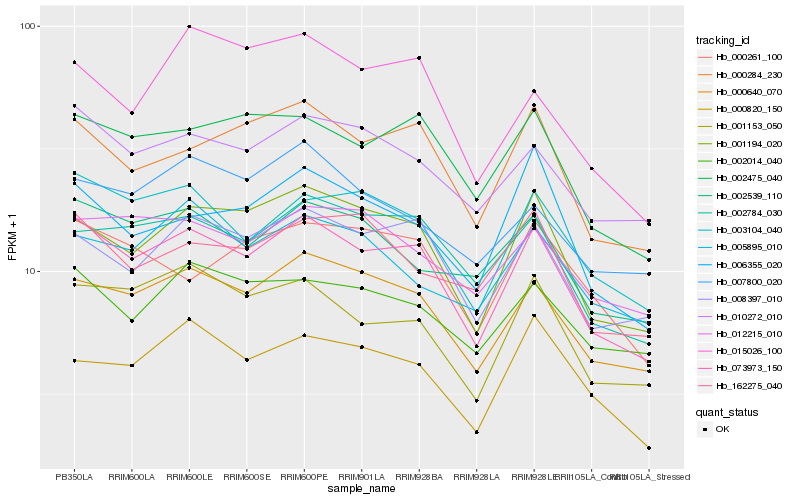
Hb_073973_150 |
0.0 |
- |
- |
PREDICTED: aminoacylase-1 isoform X1 [Jatropha curcas] |
| 2 |
Hb_000640_070 |
0.0566093341 |
- |
- |
PREDICTED: vacuolar protein sorting-associated protein 51 homolog [Jatropha curcas] |
| 3 |
Hb_001194_020 |
0.0629693934 |
- |
- |
microtubule associated protein xmap215, putative [Ricinus communis] |
| 4 |
Hb_008397_010 |
0.0676281018 |
- |
- |
PREDICTED: uncharacterized protein LOC105640192 isoform X2 [Jatropha curcas] |
| 5 |
Hb_000284_230 |
0.0680983514 |
- |
- |
PREDICTED: uncharacterized protein At5g49945-like [Jatropha curcas] |
| 6 |
Hb_003104_040 |
0.0696850894 |
- |
- |
PREDICTED: insulin-degrading enzyme isoform X1 [Jatropha curcas] |
| 7 |
Hb_002475_040 |
0.0711520405 |
- |
- |
PREDICTED: NADH dehydrogenase [ubiquinone] iron-sulfur protein 1, mitochondrial [Jatropha curcas] |
| 8 |
Hb_001153_050 |
0.073047579 |
- |
- |
PREDICTED: protein FIZZY-RELATED 2 [Jatropha curcas] |
| 9 |
Hb_002784_030 |
0.0757899148 |
- |
- |
hypothetical protein POPTR_0001s06740g [Populus trichocarpa] |
| 10 |
Hb_010272_010 |
0.0759748569 |
- |
- |
PREDICTED: endoplasmic reticulum-Golgi intermediate compartment protein 3 [Jatropha curcas] |
| 11 |
Hb_002014_040 |
0.0857125631 |
- |
- |
site-1 protease, putative [Ricinus communis] |
| 12 |
Hb_002539_110 |
0.0862927421 |
- |
- |
hypothetical protein JCGZ_15697 [Jatropha curcas] |
| 13 |
Hb_006355_020 |
0.0869665955 |
- |
- |
PREDICTED: CLIP-associated protein [Jatropha curcas] |
| 14 |
Hb_000820_150 |
0.0873972865 |
- |
- |
PREDICTED: ACT domain-containing protein ACR9-like [Jatropha curcas] |
| 15 |
Hb_012215_010 |
0.08883633 |
- |
- |
PREDICTED: YTH domain-containing family protein 1 [Populus euphratica] |
| 16 |
Hb_000261_100 |
0.091252967 |
- |
- |
PREDICTED: beta-galactosidase 9 isoform X2 [Jatropha curcas] |
| 17 |
Hb_162275_040 |
0.0926332882 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_005895_010 |
0.0941227448 |
- |
- |
PREDICTED: uncharacterized protein LOC105643387 isoform X1 [Jatropha curcas] |
| 19 |
Hb_007800_020 |
0.0942720064 |
- |
- |
PREDICTED: probable arabinosyltransferase ARAD1 isoform X1 [Jatropha curcas] |
| 20 |
Hb_015026_100 |
0.0948571629 |
- |
- |
PREDICTED: transmembrane protein 184C-like [Jatropha curcas] |