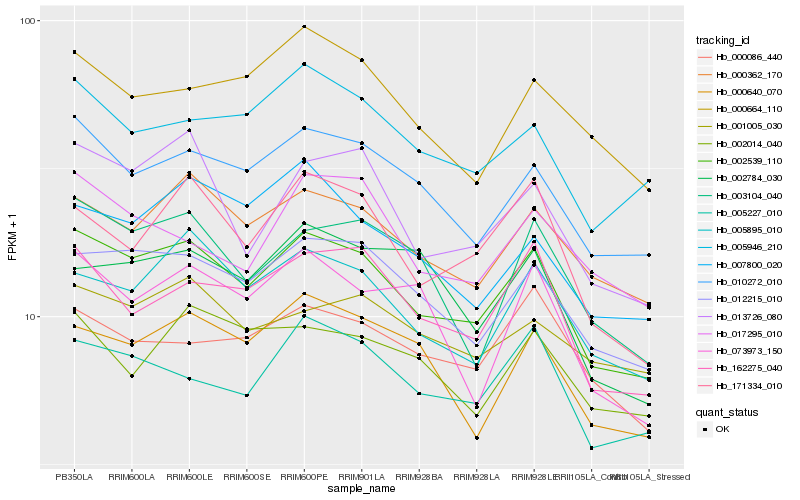
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002539_110 |
0.0 |
- |
- |
hypothetical protein JCGZ_15697 [Jatropha curcas] |
| 2 |
Hb_012215_010 |
0.0533426114 |
- |
- |
PREDICTED: YTH domain-containing family protein 1 [Populus euphratica] |
| 3 |
Hb_010272_010 |
0.0581294822 |
- |
- |
PREDICTED: endoplasmic reticulum-Golgi intermediate compartment protein 3 [Jatropha curcas] |
| 4 |
Hb_000362_170 |
0.0627563123 |
- |
- |
PREDICTED: uncharacterized protein LOC105635728 [Jatropha curcas] |
| 5 |
Hb_005895_010 |
0.0641104836 |
- |
- |
PREDICTED: uncharacterized protein LOC105643387 isoform X1 [Jatropha curcas] |
| 6 |
Hb_162275_040 |
0.0689405638 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_013726_080 |
0.0704555149 |
- |
- |
Glycogen synthase kinase-3 beta, putative [Ricinus communis] |
| 8 |
Hb_005227_010 |
0.0708364585 |
- |
- |
PREDICTED: uncharacterized protein LOC105633453 isoform X1 [Jatropha curcas] |
| 9 |
Hb_171334_010 |
0.0728617895 |
- |
- |
PREDICTED: CDPK-related kinase 5 [Jatropha curcas] |
| 10 |
Hb_000640_070 |
0.0756473029 |
- |
- |
PREDICTED: vacuolar protein sorting-associated protein 51 homolog [Jatropha curcas] |
| 11 |
Hb_005946_210 |
0.0767390594 |
- |
- |
hypothetical protein JCGZ_07228 [Jatropha curcas] |
| 12 |
Hb_003104_040 |
0.0773532875 |
- |
- |
PREDICTED: insulin-degrading enzyme isoform X1 [Jatropha curcas] |
| 13 |
Hb_007800_020 |
0.0811523324 |
- |
- |
PREDICTED: probable arabinosyltransferase ARAD1 isoform X1 [Jatropha curcas] |
| 14 |
Hb_000086_440 |
0.0813654372 |
- |
- |
PREDICTED: protein BREAST CANCER SUSCEPTIBILITY 1 homolog [Jatropha curcas] |
| 15 |
Hb_001005_030 |
0.0823054744 |
transcription factor |
TF Family: SET |
PREDICTED: histone-lysine N-methyltransferase, H3 lysine-9 specific SUVH4 isoform X3 [Jatropha curcas] |
| 16 |
Hb_000664_110 |
0.0826162569 |
- |
- |
PREDICTED: monoglyceride lipase [Jatropha curcas] |
| 17 |
Hb_002014_040 |
0.0845699666 |
- |
- |
site-1 protease, putative [Ricinus communis] |
| 18 |
Hb_017295_010 |
0.0862702269 |
- |
- |
PREDICTED: uncharacterized membrane protein At1g06890 [Jatropha curcas] |
| 19 |
Hb_073973_150 |
0.0862927421 |
- |
- |
PREDICTED: aminoacylase-1 isoform X1 [Jatropha curcas] |
| 20 |
Hb_002784_030 |
0.0876761531 |
- |
- |
hypothetical protein POPTR_0001s06740g [Populus trichocarpa] |