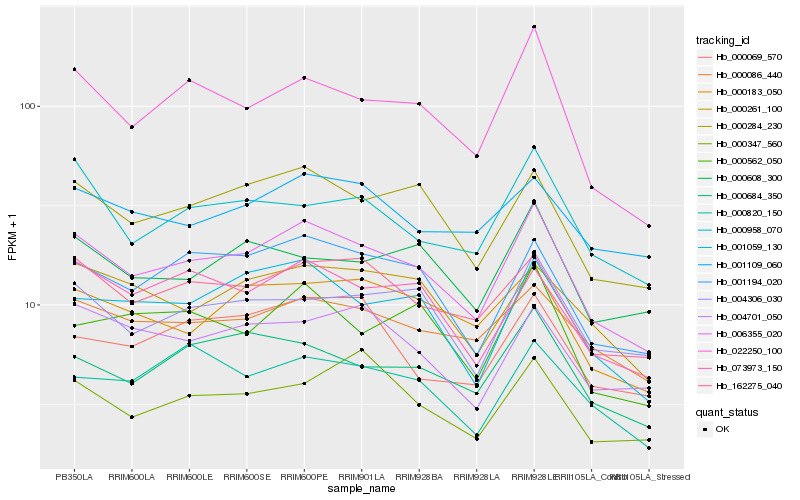
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_006355_020 |
0.0 |
- |
- |
PREDICTED: CLIP-associated protein [Jatropha curcas] |
| 2 |
Hb_000284_230 |
0.0725651736 |
- |
- |
PREDICTED: uncharacterized protein At5g49945-like [Jatropha curcas] |
| 3 |
Hb_022250_100 |
0.0726833504 |
- |
- |
calnexin, putative [Ricinus communis] |
| 4 |
Hb_162275_040 |
0.0747188879 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_001194_020 |
0.076491514 |
- |
- |
microtubule associated protein xmap215, putative [Ricinus communis] |
| 6 |
Hb_001059_130 |
0.083746722 |
- |
- |
PREDICTED: protein TRANSPORT INHIBITOR RESPONSE 1-like isoform X1 [Jatropha curcas] |
| 7 |
Hb_000086_440 |
0.0867146382 |
- |
- |
PREDICTED: protein BREAST CANCER SUSCEPTIBILITY 1 homolog [Jatropha curcas] |
| 8 |
Hb_073973_150 |
0.0869665955 |
- |
- |
PREDICTED: aminoacylase-1 isoform X1 [Jatropha curcas] |
| 9 |
Hb_004306_030 |
0.0883050835 |
- |
- |
PREDICTED: GPI transamidase component PIG-S [Jatropha curcas] |
| 10 |
Hb_000684_350 |
0.0886396246 |
- |
- |
PREDICTED: uncharacterized protein LOC105642163 isoform X1 [Jatropha curcas] |
| 11 |
Hb_000183_050 |
0.0897799748 |
- |
- |
PREDICTED: alpha-mannosidase 2 [Jatropha curcas] |
| 12 |
Hb_000261_100 |
0.0918275109 |
- |
- |
PREDICTED: beta-galactosidase 9 isoform X2 [Jatropha curcas] |
| 13 |
Hb_000958_070 |
0.0939200425 |
- |
- |
hypothetical protein PRUPE_ppa007681mg [Prunus persica] |
| 14 |
Hb_004701_050 |
0.0967185088 |
- |
- |
alpha1 family protein [Populus trichocarpa] |
| 15 |
Hb_000608_300 |
0.0979693819 |
- |
- |
PREDICTED: protein KRI1 homolog [Jatropha curcas] |
| 16 |
Hb_000562_050 |
0.0987345335 |
- |
- |
PREDICTED: uncharacterized protein LOC105635369 [Jatropha curcas] |
| 17 |
Hb_001109_060 |
0.1004014424 |
- |
- |
amino acid transporter, putative [Ricinus communis] |
| 18 |
Hb_000347_560 |
0.100813797 |
- |
- |
- |
| 19 |
Hb_000069_570 |
0.1008964361 |
- |
- |
PREDICTED: uncharacterized protein At5g05190-like isoform X1 [Jatropha curcas] |
| 20 |
Hb_000820_150 |
0.1012864428 |
- |
- |
PREDICTED: ACT domain-containing protein ACR9-like [Jatropha curcas] |