| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
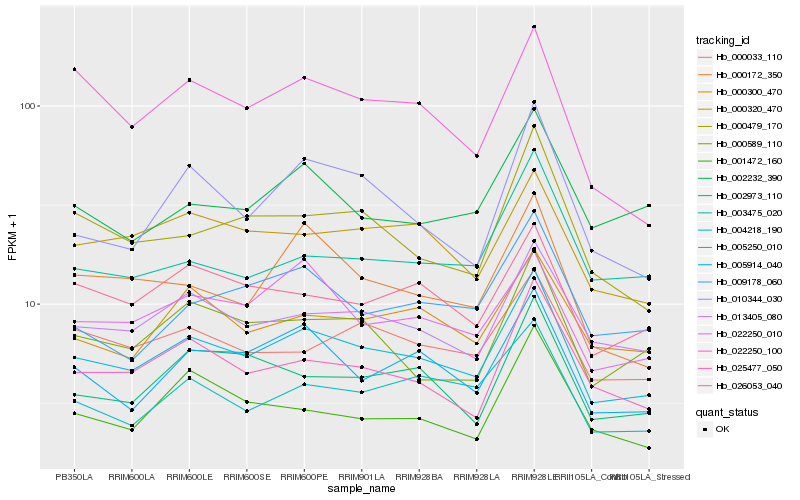
Hb_005250_010 |
0.0 |
- |
- |
hypothetical protein JCGZ_15339 [Jatropha curcas] |
| 2 |
Hb_013405_080 |
0.070147443 |
- |
- |
PREDICTED: proline--tRNA ligase [Jatropha curcas] |
| 3 |
Hb_025477_050 |
0.0820132379 |
- |
- |
ABC transporter family protein [Hevea brasiliensis] |
| 4 |
Hb_005914_040 |
0.083298678 |
- |
- |
o-linked n-acetylglucosamine transferase, ogt, putative [Ricinus communis] |
| 5 |
Hb_004218_190 |
0.0857050309 |
transcription factor |
TF Family: SNF2 |
PREDICTED: F-box protein At3g54460 isoform X1 [Jatropha curcas] |
| 6 |
Hb_026053_040 |
0.0872791554 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_000033_110 |
0.0875942728 |
- |
- |
hypothetical protein JCGZ_19253 [Jatropha curcas] |
| 8 |
Hb_000320_470 |
0.0876018713 |
- |
- |
calcium-dependent protein kinase [Hevea brasiliensis] |
| 9 |
Hb_000300_470 |
0.0885474786 |
- |
- |
PREDICTED: nucleolin-like [Jatropha curcas] |
| 10 |
Hb_000479_170 |
0.0887905984 |
- |
- |
chaperone clpb, putative [Ricinus communis] |
| 11 |
Hb_002973_110 |
0.0918630873 |
- |
- |
ABC transporter family protein [Hevea brasiliensis] |
| 12 |
Hb_022250_010 |
0.0920209912 |
- |
- |
PREDICTED: probable beta-1,4-xylosyltransferase IRX10L [Jatropha curcas] |
| 13 |
Hb_000589_110 |
0.0921182215 |
- |
- |
PREDICTED: uncharacterized protein LOC105647614 isoform X1 [Jatropha curcas] |
| 14 |
Hb_010344_030 |
0.0926404953 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_009178_060 |
0.0950776132 |
- |
- |
PREDICTED: uncharacterized aarF domain-containing protein kinase At4g31390, chloroplastic isoform X1 [Jatropha curcas] |
| 16 |
Hb_000172_350 |
0.0954384283 |
- |
- |
PREDICTED: uncharacterized protein LOC105650963 [Jatropha curcas] |
| 17 |
Hb_002232_390 |
0.0973892255 |
- |
- |
PREDICTED: uncharacterized protein LOC105635994 [Jatropha curcas] |
| 18 |
Hb_003475_020 |
0.0985160204 |
- |
- |
hypothetical protein JCGZ_13177 [Jatropha curcas] |
| 19 |
Hb_022250_100 |
0.0999086662 |
- |
- |
calnexin, putative [Ricinus communis] |
| 20 |
Hb_001472_160 |
0.1006200204 |
- |
- |
PREDICTED: mitogen-activated protein kinase kinase kinase YODA [Jatropha curcas] |