| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
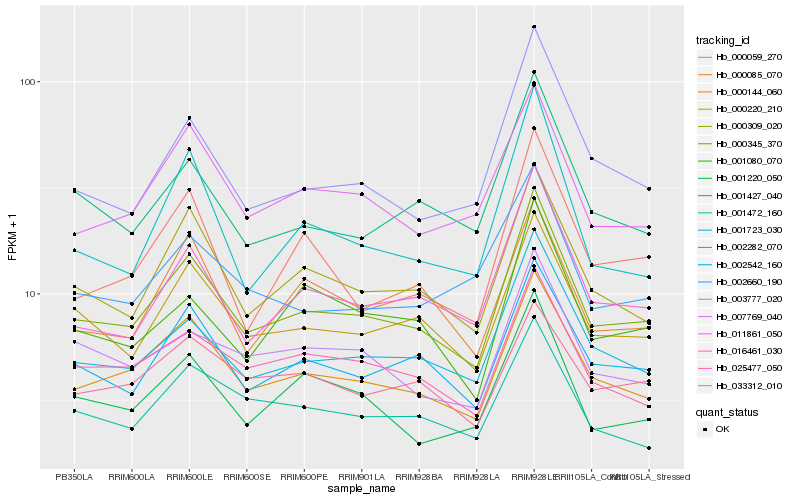
Hb_000345_370 |
0.0 |
- |
- |
PREDICTED: probable Xaa-Pro aminopeptidase P [Jatropha curcas] |
| 2 |
Hb_000144_060 |
0.0531980875 |
- |
- |
PREDICTED: uncharacterized protein At5g03900, chloroplastic isoform X1 [Jatropha curcas] |
| 3 |
Hb_002282_070 |
0.0640764716 |
- |
- |
PREDICTED: protein DJ-1 homolog C isoform X2 [Jatropha curcas] |
| 4 |
Hb_002542_160 |
0.0644099341 |
- |
- |
PREDICTED: peptide chain release factor APG3, chloroplastic [Jatropha curcas] |
| 5 |
Hb_000220_210 |
0.0647875512 |
- |
- |
PREDICTED: sec-independent protein translocase protein TATC, chloroplastic [Jatropha curcas] |
| 6 |
Hb_000309_020 |
0.0663271226 |
- |
- |
PREDICTED: uncharacterized protein LOC105641764 [Jatropha curcas] |
| 7 |
Hb_016461_030 |
0.0713901337 |
- |
- |
PREDICTED: putative leucine--tRNA ligase, mitochondrial isoform X1 [Jatropha curcas] |
| 8 |
Hb_025477_050 |
0.0797488596 |
- |
- |
ABC transporter family protein [Hevea brasiliensis] |
| 9 |
Hb_011861_050 |
0.0819135493 |
- |
- |
PREDICTED: COBW domain-containing protein 1 [Jatropha curcas] |
| 10 |
Hb_003777_020 |
0.0823556093 |
- |
- |
Fructose-1,6-bisphosphatase, cytosolic, putative [Ricinus communis] |
| 11 |
Hb_001427_040 |
0.0836939789 |
- |
- |
PREDICTED: uncharacterized protein LOC105637784 [Jatropha curcas] |
| 12 |
Hb_007769_040 |
0.0869879426 |
- |
- |
PREDICTED: probable threonine--tRNA ligase, cytoplasmic [Jatropha curcas] |
| 13 |
Hb_001472_160 |
0.0875805134 |
- |
- |
PREDICTED: mitogen-activated protein kinase kinase kinase YODA [Jatropha curcas] |
| 14 |
Hb_001220_050 |
0.090049017 |
- |
- |
DNA-damage-inducible protein f, putative [Ricinus communis] |
| 15 |
Hb_001080_070 |
0.0939851176 |
- |
- |
1,4-alpha-glucan branching enzyme [Manihot esculenta] |
| 16 |
Hb_001723_030 |
0.0957082998 |
- |
- |
PREDICTED: 30S ribosomal protein S1, chloroplastic [Jatropha curcas] |
| 17 |
Hb_033312_010 |
0.0962530215 |
- |
- |
PREDICTED: valine--tRNA ligase, mitochondrial isoform X1 [Jatropha curcas] |
| 18 |
Hb_000085_070 |
0.0992898842 |
- |
- |
PREDICTED: trigger factor-like protein TIG, Chloroplastic isoform X1 [Jatropha curcas] |
| 19 |
Hb_000059_270 |
0.0995385123 |
- |
- |
superoxide dismutase [Fe], chloroplastic [Jatropha curcas] |
| 20 |
Hb_002660_190 |
0.0997357994 |
- |
- |
PREDICTED: dnaJ protein P58IPK homolog [Jatropha curcas] |