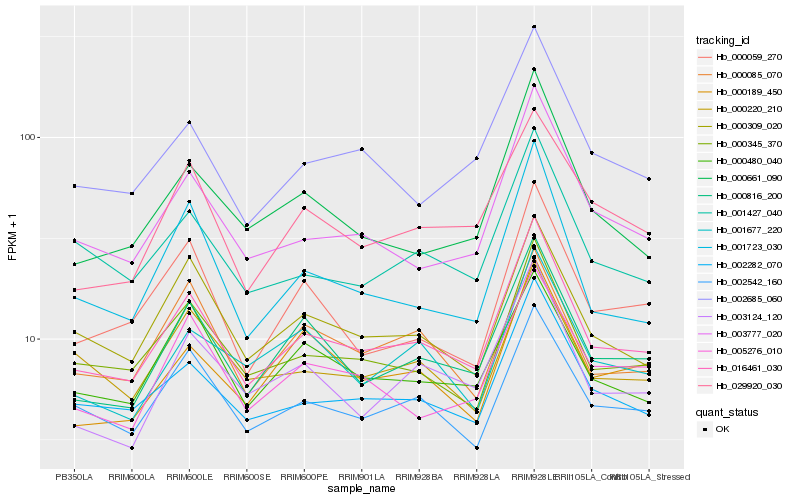
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_016461_030 |
0.0 |
- |
- |
PREDICTED: putative leucine--tRNA ligase, mitochondrial isoform X1 [Jatropha curcas] |
| 2 |
Hb_002282_070 |
0.0612862576 |
- |
- |
PREDICTED: protein DJ-1 homolog C isoform X2 [Jatropha curcas] |
| 3 |
Hb_000816_200 |
0.067480831 |
- |
- |
2-deoxyglucose-6-phosphate phosphatase, putative [Ricinus communis] |
| 4 |
Hb_000345_370 |
0.0713901337 |
- |
- |
PREDICTED: probable Xaa-Pro aminopeptidase P [Jatropha curcas] |
| 5 |
Hb_003777_020 |
0.0745819103 |
- |
- |
Fructose-1,6-bisphosphatase, cytosolic, putative [Ricinus communis] |
| 6 |
Hb_002542_160 |
0.076460601 |
- |
- |
PREDICTED: peptide chain release factor APG3, chloroplastic [Jatropha curcas] |
| 7 |
Hb_001427_040 |
0.0813955613 |
- |
- |
PREDICTED: uncharacterized protein LOC105637784 [Jatropha curcas] |
| 8 |
Hb_000309_020 |
0.082031736 |
- |
- |
PREDICTED: uncharacterized protein LOC105641764 [Jatropha curcas] |
| 9 |
Hb_002685_060 |
0.0826517124 |
- |
- |
30S ribosomal protein S5, putative [Ricinus communis] |
| 10 |
Hb_003124_120 |
0.0831839452 |
- |
- |
PREDICTED: fructose-1,6-bisphosphatase, cytosolic [Jatropha curcas] |
| 11 |
Hb_029920_030 |
0.084758236 |
- |
- |
PREDICTED: 2-Cys peroxiredoxin BAS1, chloroplastic-like [Jatropha curcas] |
| 12 |
Hb_000661_090 |
0.0880673021 |
- |
- |
PREDICTED: uncharacterized aarF domain-containing protein kinase At1g79600, chloroplastic [Jatropha curcas] |
| 13 |
Hb_001723_030 |
0.0885514149 |
- |
- |
PREDICTED: 30S ribosomal protein S1, chloroplastic [Jatropha curcas] |
| 14 |
Hb_005276_010 |
0.0888469153 |
- |
- |
hypothetical protein CICLE_v10021605mg [Citrus clementina] |
| 15 |
Hb_000480_040 |
0.0914606657 |
- |
- |
PREDICTED: L-Ala-D/L-amino acid epimerase isoform X2 [Jatropha curcas] |
| 16 |
Hb_000189_450 |
0.0932768744 |
- |
- |
Zeta-carotene desaturase, chloroplast precursor, putative [Ricinus communis] |
| 17 |
Hb_001677_220 |
0.0941371121 |
- |
- |
PREDICTED: folylpolyglutamate synthase [Jatropha curcas] |
| 18 |
Hb_000059_270 |
0.0948831235 |
- |
- |
superoxide dismutase [Fe], chloroplastic [Jatropha curcas] |
| 19 |
Hb_000085_070 |
0.0956936743 |
- |
- |
PREDICTED: trigger factor-like protein TIG, Chloroplastic isoform X1 [Jatropha curcas] |
| 20 |
Hb_000220_210 |
0.0960122104 |
- |
- |
PREDICTED: sec-independent protein translocase protein TATC, chloroplastic [Jatropha curcas] |