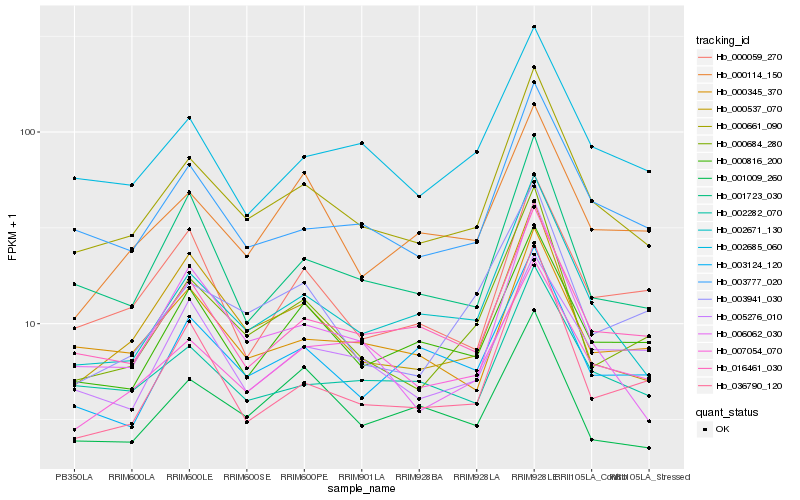
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000661_090 |
0.0 |
- |
- |
PREDICTED: uncharacterized aarF domain-containing protein kinase At1g79600, chloroplastic [Jatropha curcas] |
| 2 |
Hb_002671_130 |
0.066182679 |
- |
- |
GTP-binding protein typa/bipa, putative [Ricinus communis] |
| 3 |
Hb_003777_020 |
0.0789102405 |
- |
- |
Fructose-1,6-bisphosphatase, cytosolic, putative [Ricinus communis] |
| 4 |
Hb_016461_030 |
0.0880673021 |
- |
- |
PREDICTED: putative leucine--tRNA ligase, mitochondrial isoform X1 [Jatropha curcas] |
| 5 |
Hb_000684_280 |
0.0927937354 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At3g09650, chloroplastic [Jatropha curcas] |
| 6 |
Hb_002282_070 |
0.0959971643 |
- |
- |
PREDICTED: protein DJ-1 homolog C isoform X2 [Jatropha curcas] |
| 7 |
Hb_000537_070 |
0.0964121252 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At5g25630 [Jatropha curcas] |
| 8 |
Hb_002685_060 |
0.0982621747 |
- |
- |
30S ribosomal protein S5, putative [Ricinus communis] |
| 9 |
Hb_000816_200 |
0.1002898104 |
- |
- |
2-deoxyglucose-6-phosphate phosphatase, putative [Ricinus communis] |
| 10 |
Hb_001723_030 |
0.1010251674 |
- |
- |
PREDICTED: 30S ribosomal protein S1, chloroplastic [Jatropha curcas] |
| 11 |
Hb_006062_030 |
0.1028740004 |
- |
- |
PREDICTED: translation initiation factor IF-2, chloroplastic [Jatropha curcas] |
| 12 |
Hb_000345_370 |
0.1082051975 |
- |
- |
PREDICTED: probable Xaa-Pro aminopeptidase P [Jatropha curcas] |
| 13 |
Hb_003124_120 |
0.1091742323 |
- |
- |
PREDICTED: fructose-1,6-bisphosphatase, cytosolic [Jatropha curcas] |
| 14 |
Hb_036790_120 |
0.1100820548 |
- |
- |
PREDICTED: uncharacterized protein LOC105645122 isoform X1 [Jatropha curcas] |
| 15 |
Hb_007054_070 |
0.1116886564 |
- |
- |
PREDICTED: probable monodehydroascorbate reductase, cytoplasmic isoform 2 [Jatropha curcas] |
| 16 |
Hb_000114_150 |
0.1133496716 |
rubber biosynthesis |
Gene Name: Geranyl geranyl diphosphate synthase |
geranylgeranyl-diphosphate synthase [Hevea brasiliensis] |
| 17 |
Hb_003941_030 |
0.1134922577 |
- |
- |
Protease ecfE, putative [Ricinus communis] |
| 18 |
Hb_000059_270 |
0.1158010877 |
- |
- |
superoxide dismutase [Fe], chloroplastic [Jatropha curcas] |
| 19 |
Hb_001009_260 |
0.1171335494 |
- |
- |
PREDICTED: uncharacterized aarF domain-containing protein kinase 1 isoform X1 [Jatropha curcas] |
| 20 |
Hb_005276_010 |
0.1177028989 |
- |
- |
hypothetical protein CICLE_v10021605mg [Citrus clementina] |