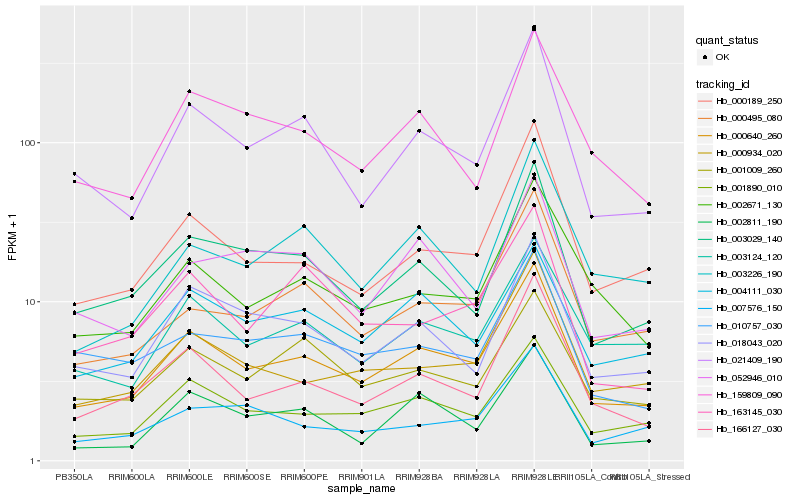
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000640_260 |
0.0 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At2g31400, chloroplastic [Jatropha curcas] |
| 2 |
Hb_001890_010 |
0.0780528268 |
- |
- |
PREDICTED: clustered mitochondria protein homolog [Jatropha curcas] |
| 3 |
Hb_021409_190 |
0.0853319884 |
- |
- |
PREDICTED: luminal-binding protein 5-like [Jatropha curcas] |
| 4 |
Hb_166127_030 |
0.089573767 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 5 |
Hb_000934_020 |
0.0991238148 |
- |
- |
PREDICTED: uncharacterized protein LOC105629153 [Jatropha curcas] |
| 6 |
Hb_000189_250 |
0.1005134099 |
- |
- |
PREDICTED: probable ribose-5-phosphate isomerase 3, chloroplastic [Jatropha curcas] |
| 7 |
Hb_003029_140 |
0.1006718576 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At2g31400, chloroplastic [Jatropha curcas] |
| 8 |
Hb_018043_020 |
0.1046425802 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein MRL1, chloroplastic [Jatropha curcas] |
| 9 |
Hb_002671_130 |
0.1048398612 |
- |
- |
GTP-binding protein typa/bipa, putative [Ricinus communis] |
| 10 |
Hb_003124_120 |
0.1049313242 |
- |
- |
PREDICTED: fructose-1,6-bisphosphatase, cytosolic [Jatropha curcas] |
| 11 |
Hb_000495_080 |
0.1096760404 |
- |
- |
Pentatricopeptide repeat (PPR-like) superfamily protein [Theobroma cacao] |
| 12 |
Hb_002811_190 |
0.1105482192 |
- |
- |
tRNA pseudouridine synthase d, putative [Ricinus communis] |
| 13 |
Hb_001009_260 |
0.1130640281 |
- |
- |
PREDICTED: uncharacterized aarF domain-containing protein kinase 1 isoform X1 [Jatropha curcas] |
| 14 |
Hb_004111_030 |
0.1134040048 |
- |
- |
PREDICTED: DNA gyrase subunit B, chloroplastic/mitochondrial-like [Jatropha curcas] |
| 15 |
Hb_159809_090 |
0.1142348696 |
rubber biosynthesis |
Gene Name: 4-hydroxy-3-methylbut-2-enyl diphosphate reductase |
4-hydroxy-3-methylbut-2-enyl diphosphate reductase [Hevea brasiliensis] |
| 16 |
Hb_010757_030 |
0.1183225143 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 17 |
Hb_007576_150 |
0.1190923246 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At3g57430, chloroplastic [Jatropha curcas] |
| 18 |
Hb_163145_030 |
0.1213972435 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At5g46580, chloroplastic [Jatropha curcas] |
| 19 |
Hb_052946_010 |
0.1229306259 |
- |
- |
PREDICTED: uncharacterized protein LOC105639103 [Jatropha curcas] |
| 20 |
Hb_003226_190 |
0.1239660173 |
- |
- |
PREDICTED: blue-light photoreceptor PHR2 [Jatropha curcas] |