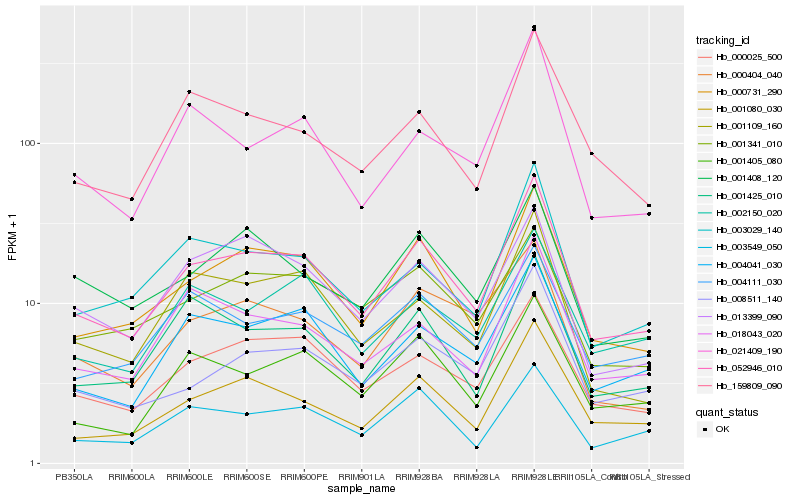
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_052946_010 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105639103 [Jatropha curcas] |
| 2 |
Hb_000731_290 |
0.0620900566 |
- |
- |
ABC transporter family protein [Hevea brasiliensis] |
| 3 |
Hb_001080_030 |
0.0844650601 |
- |
- |
AMP dependent ligase, putative [Ricinus communis] |
| 4 |
Hb_000025_500 |
0.0848994351 |
- |
- |
Ran GTPase binding protein, putative [Ricinus communis] |
| 5 |
Hb_004041_030 |
0.0911532364 |
- |
- |
PREDICTED: mitochondrial substrate carrier family protein C-like isoform X1 [Populus euphratica] |
| 6 |
Hb_008511_140 |
0.0967731104 |
transcription factor |
TF Family: mTERF |
PREDICTED: uncharacterized protein LOC105632314 [Jatropha curcas] |
| 7 |
Hb_003029_140 |
0.0968811105 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At2g31400, chloroplastic [Jatropha curcas] |
| 8 |
Hb_001405_080 |
0.1007287633 |
- |
- |
PREDICTED: acylamino-acid-releasing enzyme [Jatropha curcas] |
| 9 |
Hb_003549_050 |
0.1038627222 |
- |
- |
PREDICTED: zinc phosphodiesterase ELAC protein 2-like [Jatropha curcas] |
| 10 |
Hb_001425_010 |
0.1050621824 |
- |
- |
sugar transporter, putative [Ricinus communis] |
| 11 |
Hb_018043_020 |
0.1056567393 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein MRL1, chloroplastic [Jatropha curcas] |
| 12 |
Hb_159809_090 |
0.1067746314 |
rubber biosynthesis |
Gene Name: 4-hydroxy-3-methylbut-2-enyl diphosphate reductase |
4-hydroxy-3-methylbut-2-enyl diphosphate reductase [Hevea brasiliensis] |
| 13 |
Hb_000404_040 |
0.1083304634 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_001408_120 |
0.1091132526 |
- |
- |
dead box ATP-dependent RNA helicase, putative [Ricinus communis] |
| 15 |
Hb_002150_020 |
0.1095164494 |
- |
- |
PREDICTED: translation initiation factor IF-2, mitochondrial isoform X1 [Jatropha curcas] |
| 16 |
Hb_001109_160 |
0.1097662144 |
- |
- |
DNA binding protein, putative [Ricinus communis] |
| 17 |
Hb_021409_190 |
0.1111506955 |
- |
- |
PREDICTED: luminal-binding protein 5-like [Jatropha curcas] |
| 18 |
Hb_004111_030 |
0.1111595434 |
- |
- |
PREDICTED: DNA gyrase subunit B, chloroplastic/mitochondrial-like [Jatropha curcas] |
| 19 |
Hb_013399_090 |
0.1123775071 |
- |
- |
PREDICTED: phospholipid:diacylglycerol acyltransferase 1-like [Jatropha curcas] |
| 20 |
Hb_001341_010 |
0.112595824 |
- |
- |
PREDICTED: carbamoyl-phosphate synthase large chain, chloroplastic [Jatropha curcas] |