| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
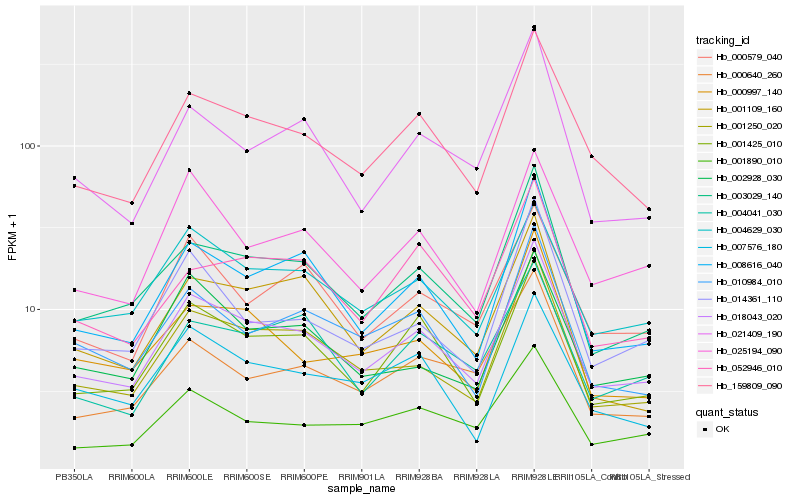
Hb_018043_020 |
0.0 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein MRL1, chloroplastic [Jatropha curcas] |
| 2 |
Hb_003029_140 |
0.0640337029 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At2g31400, chloroplastic [Jatropha curcas] |
| 3 |
Hb_159809_090 |
0.0640517181 |
rubber biosynthesis |
Gene Name: 4-hydroxy-3-methylbut-2-enyl diphosphate reductase |
4-hydroxy-3-methylbut-2-enyl diphosphate reductase [Hevea brasiliensis] |
| 4 |
Hb_001250_020 |
0.0687169959 |
- |
- |
PREDICTED: CRS2-associated factor 2, chloroplastic [Jatropha curcas] |
| 5 |
Hb_008616_040 |
0.0713503786 |
- |
- |
PREDICTED: RNA-binding protein CP33, chloroplastic [Jatropha curcas] |
| 6 |
Hb_001425_010 |
0.0739736219 |
- |
- |
sugar transporter, putative [Ricinus communis] |
| 7 |
Hb_004629_030 |
0.0883701579 |
- |
- |
ABC transporter family protein [Hevea brasiliensis] |
| 8 |
Hb_010984_010 |
0.0939392183 |
- |
- |
PREDICTED: translation factor GUF1 homolog, chloroplastic isoform X1 [Jatropha curcas] |
| 9 |
Hb_001890_010 |
0.0963695058 |
- |
- |
PREDICTED: clustered mitochondria protein homolog [Jatropha curcas] |
| 10 |
Hb_021409_190 |
0.0965310768 |
- |
- |
PREDICTED: luminal-binding protein 5-like [Jatropha curcas] |
| 11 |
Hb_000997_140 |
0.0987449022 |
- |
- |
PREDICTED: chloroplastic group IIA intron splicing facilitator CRS1, chloroplastic [Jatropha curcas] |
| 12 |
Hb_014361_110 |
0.0999952862 |
- |
- |
RNA binding protein, putative [Ricinus communis] |
| 13 |
Hb_000579_040 |
0.1001461515 |
- |
- |
PREDICTED: putative GTP-binding protein 6 [Jatropha curcas] |
| 14 |
Hb_025194_090 |
0.101323056 |
- |
- |
coproporphyrinogen III oxidase, putative [Ricinus communis] |
| 15 |
Hb_002928_030 |
0.102321128 |
- |
- |
hypothetical protein CICLE_v10001788mg [Citrus clementina] |
| 16 |
Hb_004041_030 |
0.1032190448 |
- |
- |
PREDICTED: mitochondrial substrate carrier family protein C-like isoform X1 [Populus euphratica] |
| 17 |
Hb_007576_180 |
0.10385253 |
- |
- |
PREDICTED: probable protein phosphatase 2C 62 isoform X5 [Jatropha curcas] |
| 18 |
Hb_000640_260 |
0.1046425802 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At2g31400, chloroplastic [Jatropha curcas] |
| 19 |
Hb_001109_160 |
0.104668351 |
- |
- |
DNA binding protein, putative [Ricinus communis] |
| 20 |
Hb_052946_010 |
0.1056567393 |
- |
- |
PREDICTED: uncharacterized protein LOC105639103 [Jatropha curcas] |