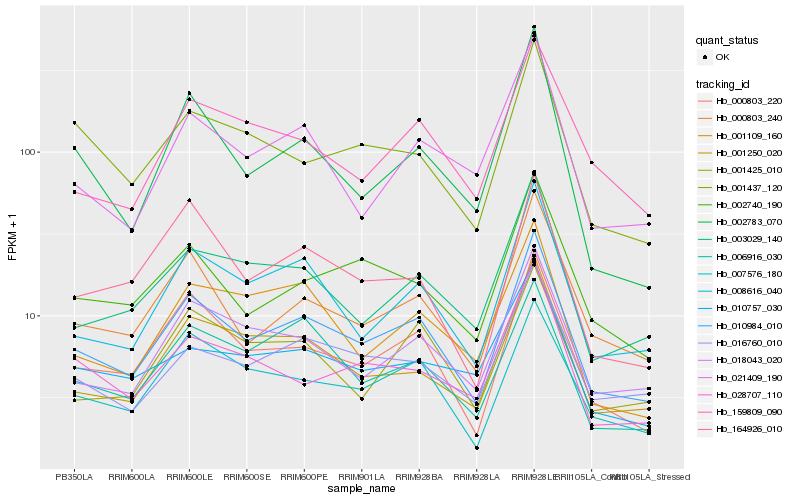
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_010984_010 |
0.0 |
- |
- |
PREDICTED: translation factor GUF1 homolog, chloroplastic isoform X1 [Jatropha curcas] |
| 2 |
Hb_000803_220 |
0.0776817539 |
- |
- |
PREDICTED: magnesium-chelatase subunit ChlD, chloroplastic [Jatropha curcas] |
| 3 |
Hb_008616_040 |
0.0855662724 |
- |
- |
PREDICTED: RNA-binding protein CP33, chloroplastic [Jatropha curcas] |
| 4 |
Hb_000803_240 |
0.088596881 |
- |
- |
hypothetical protein CICLE_v100143612mg, partial [Citrus clementina] |
| 5 |
Hb_007576_180 |
0.0892716948 |
- |
- |
PREDICTED: probable protein phosphatase 2C 62 isoform X5 [Jatropha curcas] |
| 6 |
Hb_018043_020 |
0.0939392183 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein MRL1, chloroplastic [Jatropha curcas] |
| 7 |
Hb_016760_010 |
0.0952467651 |
- |
- |
ABC transporter family protein [Hevea brasiliensis] |
| 8 |
Hb_001250_020 |
0.0977772006 |
- |
- |
PREDICTED: CRS2-associated factor 2, chloroplastic [Jatropha curcas] |
| 9 |
Hb_003029_140 |
0.1012536264 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At2g31400, chloroplastic [Jatropha curcas] |
| 10 |
Hb_002740_190 |
0.1031673894 |
- |
- |
PREDICTED: DEAD-box ATP-dependent RNA helicase 3, chloroplastic [Jatropha curcas] |
| 11 |
Hb_006916_030 |
0.1062648862 |
- |
- |
PREDICTED: uncharacterized protein LOC105646760 [Jatropha curcas] |
| 12 |
Hb_159809_090 |
0.1097738326 |
rubber biosynthesis |
Gene Name: 4-hydroxy-3-methylbut-2-enyl diphosphate reductase |
4-hydroxy-3-methylbut-2-enyl diphosphate reductase [Hevea brasiliensis] |
| 13 |
Hb_001425_010 |
0.1098360227 |
- |
- |
sugar transporter, putative [Ricinus communis] |
| 14 |
Hb_021409_190 |
0.113369116 |
- |
- |
PREDICTED: luminal-binding protein 5-like [Jatropha curcas] |
| 15 |
Hb_001109_160 |
0.1148456231 |
- |
- |
DNA binding protein, putative [Ricinus communis] |
| 16 |
Hb_010757_030 |
0.1156963075 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 17 |
Hb_002783_070 |
0.1157582402 |
- |
- |
PREDICTED: endoplasmin homolog [Jatropha curcas] |
| 18 |
Hb_001437_120 |
0.1165491453 |
- |
- |
PREDICTED: ruBisCO large subunit-binding protein subunit alpha [Jatropha curcas] |
| 19 |
Hb_028707_110 |
0.1174226546 |
- |
- |
PREDICTED: uncharacterized protein LOC105645798 [Jatropha curcas] |
| 20 |
Hb_164926_010 |
0.1179320393 |
- |
- |
Granule-bound starch synthase 1, chloroplastic/amyloplastic [Gossypium arboreum] |