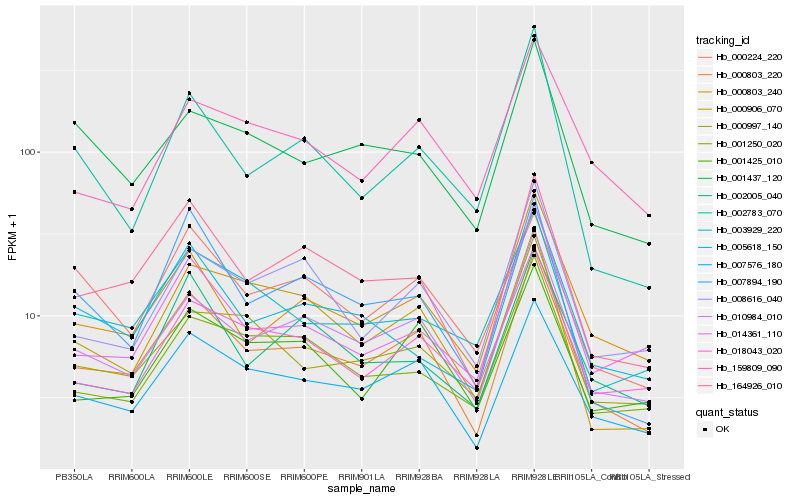
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000803_220 |
0.0 |
- |
- |
PREDICTED: magnesium-chelatase subunit ChlD, chloroplastic [Jatropha curcas] |
| 2 |
Hb_007576_180 |
0.0676437033 |
- |
- |
PREDICTED: probable protein phosphatase 2C 62 isoform X5 [Jatropha curcas] |
| 3 |
Hb_010984_010 |
0.0776817539 |
- |
- |
PREDICTED: translation factor GUF1 homolog, chloroplastic isoform X1 [Jatropha curcas] |
| 4 |
Hb_164926_010 |
0.0872288417 |
- |
- |
Granule-bound starch synthase 1, chloroplastic/amyloplastic [Gossypium arboreum] |
| 5 |
Hb_000803_240 |
0.104946921 |
- |
- |
hypothetical protein CICLE_v100143612mg, partial [Citrus clementina] |
| 6 |
Hb_001425_010 |
0.1161157671 |
- |
- |
sugar transporter, putative [Ricinus communis] |
| 7 |
Hb_018043_020 |
0.1161606625 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein MRL1, chloroplastic [Jatropha curcas] |
| 8 |
Hb_007894_190 |
0.119544768 |
- |
- |
PREDICTED: uncharacterized protein LOC105632935 [Jatropha curcas] |
| 9 |
Hb_008616_040 |
0.1213959591 |
- |
- |
PREDICTED: RNA-binding protein CP33, chloroplastic [Jatropha curcas] |
| 10 |
Hb_000906_070 |
0.1263911862 |
- |
- |
PREDICTED: protein SCAR2-like isoform X1 [Jatropha curcas] |
| 11 |
Hb_159809_090 |
0.1274729581 |
rubber biosynthesis |
Gene Name: 4-hydroxy-3-methylbut-2-enyl diphosphate reductase |
4-hydroxy-3-methylbut-2-enyl diphosphate reductase [Hevea brasiliensis] |
| 12 |
Hb_001250_020 |
0.1284047434 |
- |
- |
PREDICTED: CRS2-associated factor 2, chloroplastic [Jatropha curcas] |
| 13 |
Hb_002005_040 |
0.1284835722 |
- |
- |
PREDICTED: carboxyl-terminal-processing peptidase 1, chloroplastic isoform X2 [Jatropha curcas] |
| 14 |
Hb_003929_220 |
0.128630065 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 15 |
Hb_005618_150 |
0.1298082391 |
- |
- |
PREDICTED: protein translocase subunit SecA, chloroplastic [Jatropha curcas] |
| 16 |
Hb_014361_110 |
0.1301117483 |
- |
- |
RNA binding protein, putative [Ricinus communis] |
| 17 |
Hb_000997_140 |
0.1301975982 |
- |
- |
PREDICTED: chloroplastic group IIA intron splicing facilitator CRS1, chloroplastic [Jatropha curcas] |
| 18 |
Hb_001437_120 |
0.1317947613 |
- |
- |
PREDICTED: ruBisCO large subunit-binding protein subunit alpha [Jatropha curcas] |
| 19 |
Hb_002783_070 |
0.1318630126 |
- |
- |
PREDICTED: endoplasmin homolog [Jatropha curcas] |
| 20 |
Hb_000224_220 |
0.132167492 |
- |
- |
PREDICTED: protein disulfide isomerase-like 2-3 [Jatropha curcas] |