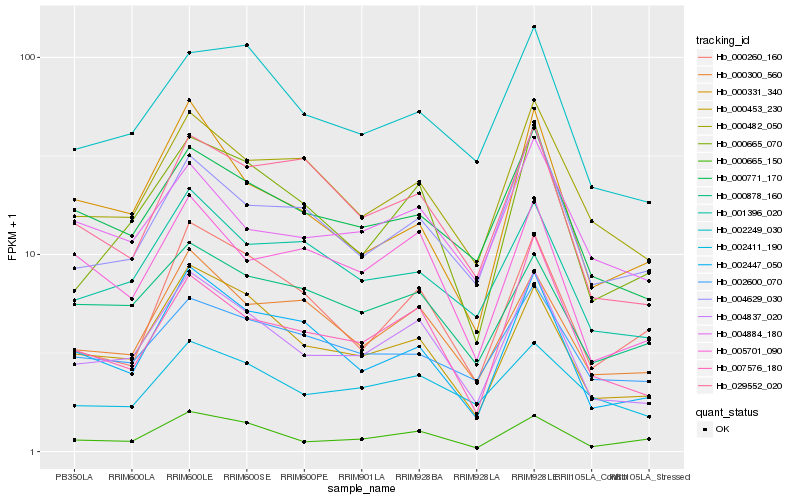
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004837_020 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105648271 [Jatropha curcas] |
| 2 |
Hb_000453_230 |
0.0794789615 |
- |
- |
PREDICTED: NO-associated protein 1, chloroplastic/mitochondrial [Jatropha curcas] |
| 3 |
Hb_000665_070 |
0.0937508591 |
transcription factor |
TF Family: HB |
PREDICTED: homeobox protein knotted-1-like 3 isoform X1 [Jatropha curcas] |
| 4 |
Hb_000771_170 |
0.0984849555 |
- |
- |
PREDICTED: ATP-dependent zinc metalloprotease FTSH 11, chloroplastic/mitochondrial [Jatropha curcas] |
| 5 |
Hb_000665_150 |
0.100509641 |
- |
- |
heparanase, putative [Ricinus communis] |
| 6 |
Hb_007576_180 |
0.1015618218 |
- |
- |
PREDICTED: probable protein phosphatase 2C 62 isoform X5 [Jatropha curcas] |
| 7 |
Hb_002447_050 |
0.103435933 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 8 |
Hb_000300_560 |
0.1042418412 |
- |
- |
PREDICTED: probable glycosyltransferase At5g03795 isoform X1 [Jatropha curcas] |
| 9 |
Hb_000482_050 |
0.1077099775 |
- |
- |
PREDICTED: UDP-sulfoquinovose synthase, chloroplastic [Jatropha curcas] |
| 10 |
Hb_002249_030 |
0.1086766908 |
- |
- |
PREDICTED: chaperone protein ClpC, chloroplastic [Jatropha curcas] |
| 11 |
Hb_001396_020 |
0.1090062897 |
- |
- |
glucose-6-phosphate dehydrogenase [Hevea brasiliensis] |
| 12 |
Hb_004629_030 |
0.1115004552 |
- |
- |
ABC transporter family protein [Hevea brasiliensis] |
| 13 |
Hb_005701_090 |
0.112094652 |
- |
- |
PREDICTED: protein ACCUMULATION AND REPLICATION OF CHLOROPLASTS 6, chloroplastic isoform X1 [Jatropha curcas] |
| 14 |
Hb_000878_160 |
0.121449384 |
- |
- |
PREDICTED: D-lactate dehydrogenase [cytochrome], mitochondrial [Jatropha curcas] |
| 15 |
Hb_002411_190 |
0.1215948 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: putative disease resistance protein RGA1 [Jatropha curcas] |
| 16 |
Hb_000331_340 |
0.1217893647 |
- |
- |
PREDICTED: uncharacterized protein LOC105640234 [Jatropha curcas] |
| 17 |
Hb_029552_020 |
0.122097884 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_000260_160 |
0.1249822997 |
transcription factor |
TF Family: GNAT |
ATP binding protein, putative [Ricinus communis] |
| 19 |
Hb_002600_070 |
0.1252732265 |
transcription factor |
TF Family: TAZ |
PREDICTED: histone acetyltransferase HAC12 [Jatropha curcas] |
| 20 |
Hb_004884_180 |
0.1263864673 |
- |
- |
PREDICTED: protein TIC 56, chloroplastic [Jatropha curcas] |