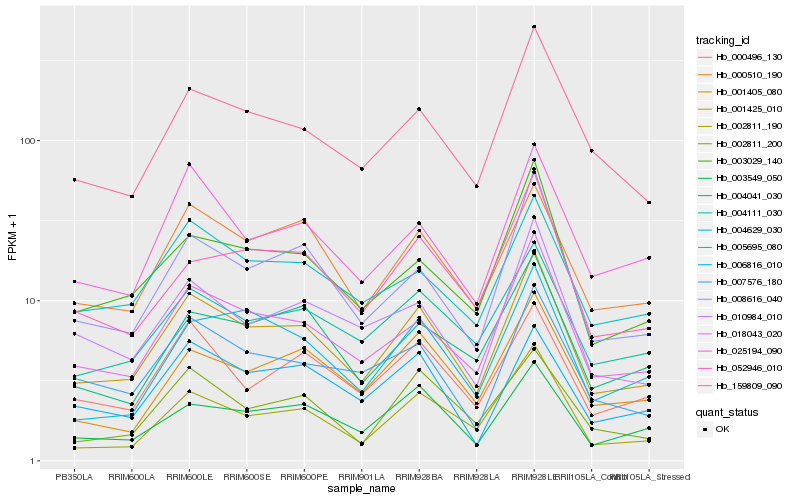
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001425_010 |
0.0 |
- |
- |
sugar transporter, putative [Ricinus communis] |
| 2 |
Hb_018043_020 |
0.0739736219 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein MRL1, chloroplastic [Jatropha curcas] |
| 3 |
Hb_008616_040 |
0.0881948463 |
- |
- |
PREDICTED: RNA-binding protein CP33, chloroplastic [Jatropha curcas] |
| 4 |
Hb_159809_090 |
0.0933330445 |
rubber biosynthesis |
Gene Name: 4-hydroxy-3-methylbut-2-enyl diphosphate reductase |
4-hydroxy-3-methylbut-2-enyl diphosphate reductase [Hevea brasiliensis] |
| 5 |
Hb_000510_190 |
0.0943010956 |
- |
- |
glutathione reductase [Hevea brasiliensis] |
| 6 |
Hb_003549_050 |
0.0971312351 |
- |
- |
PREDICTED: zinc phosphodiesterase ELAC protein 2-like [Jatropha curcas] |
| 7 |
Hb_003029_140 |
0.0977009692 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At2g31400, chloroplastic [Jatropha curcas] |
| 8 |
Hb_004111_030 |
0.0981281195 |
- |
- |
PREDICTED: DNA gyrase subunit B, chloroplastic/mitochondrial-like [Jatropha curcas] |
| 9 |
Hb_002811_190 |
0.09846697 |
- |
- |
tRNA pseudouridine synthase d, putative [Ricinus communis] |
| 10 |
Hb_004629_030 |
0.1020959218 |
- |
- |
ABC transporter family protein [Hevea brasiliensis] |
| 11 |
Hb_007576_180 |
0.1021171489 |
- |
- |
PREDICTED: probable protein phosphatase 2C 62 isoform X5 [Jatropha curcas] |
| 12 |
Hb_000496_130 |
0.1029434084 |
- |
- |
PREDICTED: fructokinase-1 [Jatropha curcas] |
| 13 |
Hb_025194_090 |
0.1049575932 |
- |
- |
coproporphyrinogen III oxidase, putative [Ricinus communis] |
| 14 |
Hb_052946_010 |
0.1050621824 |
- |
- |
PREDICTED: uncharacterized protein LOC105639103 [Jatropha curcas] |
| 15 |
Hb_006816_010 |
0.1076756401 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_001405_080 |
0.108914262 |
- |
- |
PREDICTED: acylamino-acid-releasing enzyme [Jatropha curcas] |
| 17 |
Hb_004041_030 |
0.1096678032 |
- |
- |
PREDICTED: mitochondrial substrate carrier family protein C-like isoform X1 [Populus euphratica] |
| 18 |
Hb_010984_010 |
0.1098360227 |
- |
- |
PREDICTED: translation factor GUF1 homolog, chloroplastic isoform X1 [Jatropha curcas] |
| 19 |
Hb_002811_200 |
0.1105311346 |
- |
- |
PREDICTED: protein IQ-DOMAIN 14-like [Jatropha curcas] |
| 20 |
Hb_005695_080 |
0.1108388025 |
- |
- |
PREDICTED: acetyl-CoA carboxylase 1-like [Jatropha curcas] |