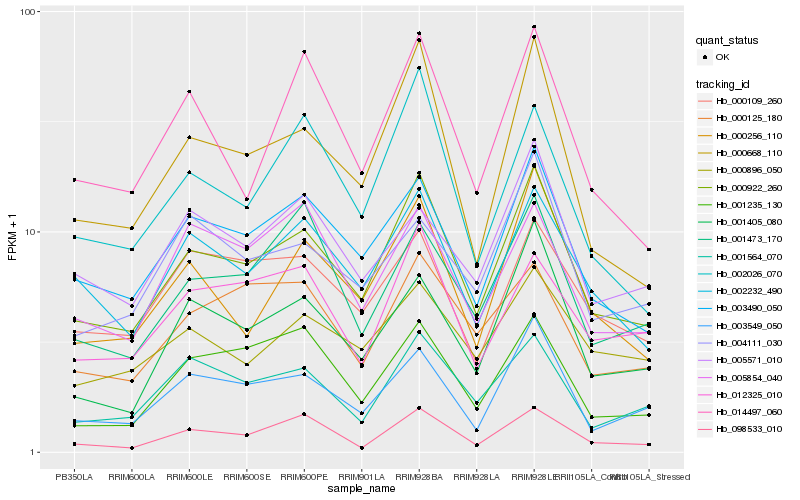
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000922_260 |
0.0 |
- |
- |
PREDICTED: pantothenate kinase 2 [Jatropha curcas] |
| 2 |
Hb_003490_050 |
0.0818486898 |
- |
- |
PREDICTED: probable cyclic nucleotide-gated ion channel 20, chloroplastic [Jatropha curcas] |
| 3 |
Hb_000668_110 |
0.0891804679 |
- |
- |
PREDICTED: acetyl-CoA carboxylase 1-like [Jatropha curcas] |
| 4 |
Hb_000256_110 |
0.0975747834 |
- |
- |
PREDICTED: aspartate aminotransferase, chloroplastic [Jatropha curcas] |
| 5 |
Hb_002232_490 |
0.0985376083 |
- |
- |
PREDICTED: probable sphingolipid transporter spinster homolog 2 isoform X1 [Jatropha curcas] |
| 6 |
Hb_003549_050 |
0.1002413964 |
- |
- |
PREDICTED: zinc phosphodiesterase ELAC protein 2-like [Jatropha curcas] |
| 7 |
Hb_001405_080 |
0.1071127624 |
- |
- |
PREDICTED: acylamino-acid-releasing enzyme [Jatropha curcas] |
| 8 |
Hb_001564_070 |
0.1077469513 |
- |
- |
PREDICTED: G patch domain-containing protein TGH [Jatropha curcas] |
| 9 |
Hb_000896_050 |
0.1117948319 |
- |
- |
PREDICTED: uncharacterized protein LOC105637668 [Jatropha curcas] |
| 10 |
Hb_002026_070 |
0.1122228916 |
- |
- |
hypothetical protein PHAVU_002G2910001g, partial [Phaseolus vulgaris] |
| 11 |
Hb_014497_060 |
0.1148511786 |
- |
- |
phosphofructokinase, putative [Ricinus communis] |
| 12 |
Hb_012325_010 |
0.1157462696 |
- |
- |
hypothetical protein RCOM_0068670 [Ricinus communis] |
| 13 |
Hb_000109_260 |
0.1168738504 |
- |
- |
PREDICTED: WAT1-related protein At3g28050-like isoform X1 [Jatropha curcas] |
| 14 |
Hb_000125_180 |
0.117922073 |
- |
- |
PREDICTED: flowering time control protein FPA isoform X1 [Jatropha curcas] |
| 15 |
Hb_098533_010 |
0.1195842191 |
- |
- |
hypothetical protein L484_003492 [Morus notabilis] |
| 16 |
Hb_001473_170 |
0.1208743671 |
- |
- |
PREDICTED: non-lysosomal glucosylceramidase [Jatropha curcas] |
| 17 |
Hb_001235_130 |
0.1214045524 |
- |
- |
- |
| 18 |
Hb_004111_030 |
0.1243930496 |
- |
- |
PREDICTED: DNA gyrase subunit B, chloroplastic/mitochondrial-like [Jatropha curcas] |
| 19 |
Hb_005571_010 |
0.1257346062 |
- |
- |
PREDICTED: 15-cis-phytoene desaturase, chloroplastic/chromoplastic [Jatropha curcas] |
| 20 |
Hb_005854_040 |
0.1260894752 |
- |
- |
conserved hypothetical protein [Ricinus communis] |