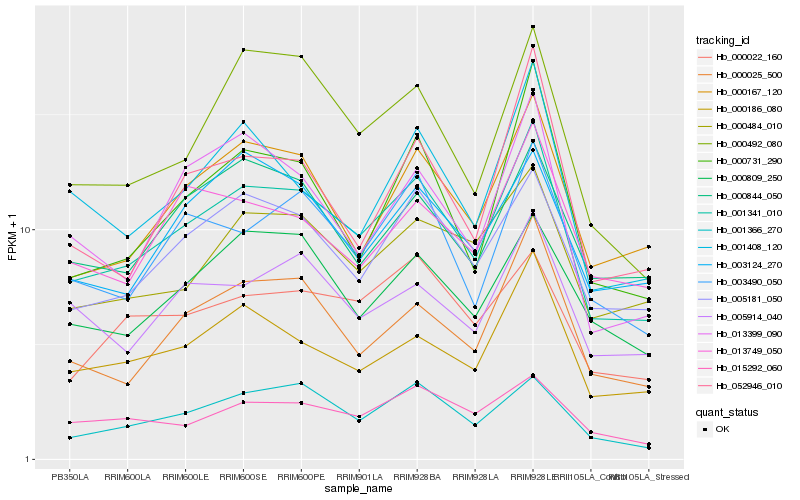
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001341_010 |
0.0 |
- |
- |
PREDICTED: carbamoyl-phosphate synthase large chain, chloroplastic [Jatropha curcas] |
| 2 |
Hb_005181_050 |
0.0850260563 |
- |
- |
PREDICTED: phytochrome B isoform X1 [Jatropha curcas] |
| 3 |
Hb_000167_120 |
0.087878315 |
- |
- |
PREDICTED: U-box domain-containing protein 14 [Jatropha curcas] |
| 4 |
Hb_000025_500 |
0.0886339416 |
- |
- |
Ran GTPase binding protein, putative [Ricinus communis] |
| 5 |
Hb_001366_270 |
0.0929898904 |
- |
- |
PREDICTED: LRR receptor-like serine/threonine-protein kinase RPK2 [Jatropha curcas] |
| 6 |
Hb_000186_080 |
0.0934762557 |
- |
- |
PREDICTED: protein unc-45 homolog B-like [Gossypium raimondii] |
| 7 |
Hb_003490_050 |
0.0943800568 |
- |
- |
PREDICTED: probable cyclic nucleotide-gated ion channel 20, chloroplastic [Jatropha curcas] |
| 8 |
Hb_000484_010 |
0.0993084576 |
- |
- |
PREDICTED: probable ubiquitin-conjugating enzyme E2 23 [Jatropha curcas] |
| 9 |
Hb_000731_290 |
0.1037646003 |
- |
- |
ABC transporter family protein [Hevea brasiliensis] |
| 10 |
Hb_000492_080 |
0.104576878 |
- |
- |
PREDICTED: probable sucrose-phosphate synthase 1 [Jatropha curcas] |
| 11 |
Hb_000809_250 |
0.1049335659 |
- |
- |
PREDICTED: uncharacterized protein LOC105639683 [Jatropha curcas] |
| 12 |
Hb_013399_090 |
0.1068811486 |
- |
- |
PREDICTED: phospholipid:diacylglycerol acyltransferase 1-like [Jatropha curcas] |
| 13 |
Hb_001408_120 |
0.1077993541 |
- |
- |
dead box ATP-dependent RNA helicase, putative [Ricinus communis] |
| 14 |
Hb_000844_050 |
0.1080023793 |
transcription factor |
TF Family: B3 |
PREDICTED: B3 domain-containing protein Os01g0723500-like [Jatropha curcas] |
| 15 |
Hb_005914_040 |
0.108285303 |
- |
- |
o-linked n-acetylglucosamine transferase, ogt, putative [Ricinus communis] |
| 16 |
Hb_000022_160 |
0.1101209183 |
- |
- |
- |
| 17 |
Hb_015292_060 |
0.1101399876 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At5g38730 [Jatropha curcas] |
| 18 |
Hb_013749_050 |
0.1108581058 |
- |
- |
PREDICTED: uncharacterized protein LOC105647765 isoform X2 [Jatropha curcas] |
| 19 |
Hb_003124_270 |
0.1124982266 |
transcription factor |
TF Family: LUG |
hypothetical protein JCGZ_13223 [Jatropha curcas] |
| 20 |
Hb_052946_010 |
0.112595824 |
- |
- |
PREDICTED: uncharacterized protein LOC105639103 [Jatropha curcas] |