| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
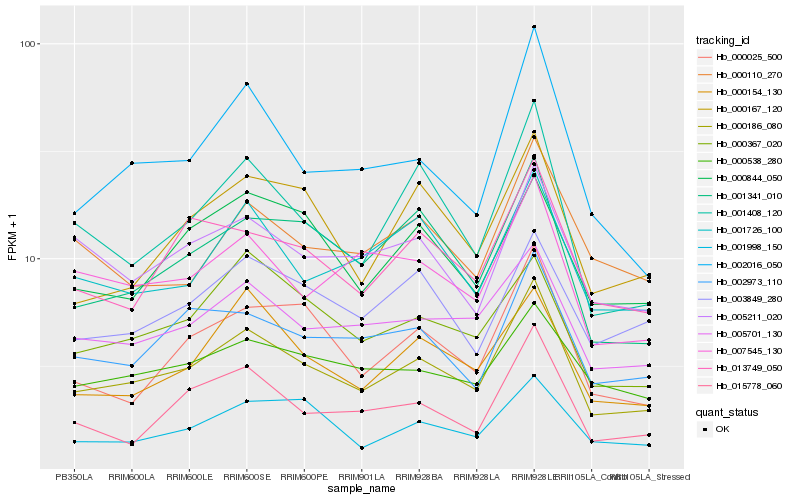
Hb_000186_080 |
0.0 |
- |
- |
PREDICTED: protein unc-45 homolog B-like [Gossypium raimondii] |
| 2 |
Hb_005701_130 |
0.0892220054 |
transcription factor |
TF Family: SWI/SNF-SWI3 |
PREDICTED: lysine-specific histone demethylase 1 homolog 3 [Jatropha curcas] |
| 3 |
Hb_001408_120 |
0.0916902143 |
- |
- |
dead box ATP-dependent RNA helicase, putative [Ricinus communis] |
| 4 |
Hb_001341_010 |
0.0934762557 |
- |
- |
PREDICTED: carbamoyl-phosphate synthase large chain, chloroplastic [Jatropha curcas] |
| 5 |
Hb_002016_050 |
0.094516578 |
- |
- |
PREDICTED: translocase of chloroplast 159, chloroplastic [Jatropha curcas] |
| 6 |
Hb_013749_050 |
0.0947165266 |
- |
- |
PREDICTED: uncharacterized protein LOC105647765 isoform X2 [Jatropha curcas] |
| 7 |
Hb_002973_110 |
0.0952044504 |
- |
- |
ABC transporter family protein [Hevea brasiliensis] |
| 8 |
Hb_000025_500 |
0.1016436763 |
- |
- |
Ran GTPase binding protein, putative [Ricinus communis] |
| 9 |
Hb_015778_060 |
0.1021370433 |
- |
- |
PREDICTED: uncharacterized protein LOC105650696 [Jatropha curcas] |
| 10 |
Hb_007545_130 |
0.1036091949 |
- |
- |
PREDICTED: zinc finger protein VAR3, chloroplastic [Jatropha curcas] |
| 11 |
Hb_001726_100 |
0.1040139658 |
transcription factor |
TF Family: C3H |
PREDICTED: zinc finger CCCH domain-containing protein 19 isoform X2 [Jatropha curcas] |
| 12 |
Hb_000154_130 |
0.1041146654 |
- |
- |
PREDICTED: flowering time control protein FPA isoform X1 [Jatropha curcas] |
| 13 |
Hb_005211_020 |
0.1046236218 |
- |
- |
PREDICTED: serine/threonine-protein kinase prpf4B-like isoform X1 [Jatropha curcas] |
| 14 |
Hb_001998_150 |
0.10509526 |
- |
- |
hypothetical protein JCGZ_09140 [Jatropha curcas] |
| 15 |
Hb_000367_020 |
0.1072876542 |
- |
- |
PREDICTED: uncharacterized protein LOC105640608 [Jatropha curcas] |
| 16 |
Hb_000167_120 |
0.107790265 |
- |
- |
PREDICTED: U-box domain-containing protein 14 [Jatropha curcas] |
| 17 |
Hb_000538_280 |
0.107868976 |
- |
- |
PREDICTED: uncharacterized protein LOC105642112 [Jatropha curcas] |
| 18 |
Hb_003849_280 |
0.1090673922 |
- |
- |
PREDICTED: uncharacterized protein LOC105644977 [Jatropha curcas] |
| 19 |
Hb_000844_050 |
0.109773522 |
transcription factor |
TF Family: B3 |
PREDICTED: B3 domain-containing protein Os01g0723500-like [Jatropha curcas] |
| 20 |
Hb_000110_270 |
0.1122613442 |
- |
- |
prp4, putative [Ricinus communis] |