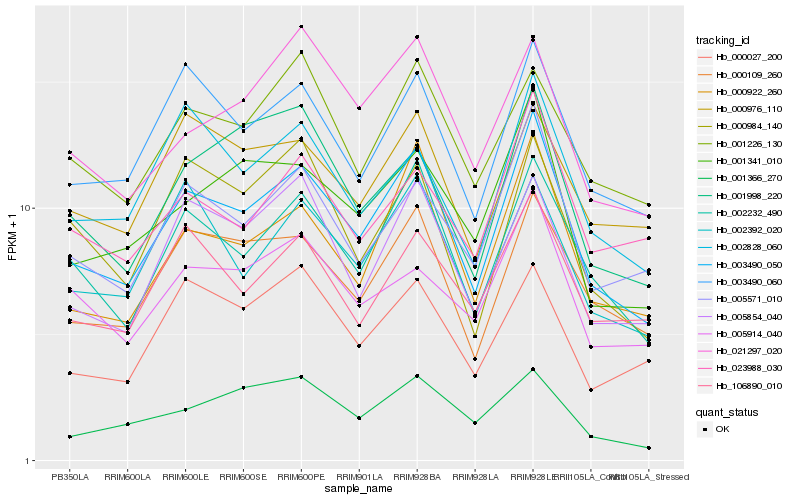
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003490_050 |
0.0 |
- |
- |
PREDICTED: probable cyclic nucleotide-gated ion channel 20, chloroplastic [Jatropha curcas] |
| 2 |
Hb_002232_490 |
0.0724703333 |
- |
- |
PREDICTED: probable sphingolipid transporter spinster homolog 2 isoform X1 [Jatropha curcas] |
| 3 |
Hb_000922_260 |
0.0818486898 |
- |
- |
PREDICTED: pantothenate kinase 2 [Jatropha curcas] |
| 4 |
Hb_003490_060 |
0.0866377002 |
- |
- |
PREDICTED: probable cyclic nucleotide-gated ion channel 20, chloroplastic isoform X2 [Sesamum indicum] |
| 5 |
Hb_000109_260 |
0.0866576689 |
- |
- |
PREDICTED: WAT1-related protein At3g28050-like isoform X1 [Jatropha curcas] |
| 6 |
Hb_005571_010 |
0.0887799299 |
- |
- |
PREDICTED: 15-cis-phytoene desaturase, chloroplastic/chromoplastic [Jatropha curcas] |
| 7 |
Hb_001341_010 |
0.0943800568 |
- |
- |
PREDICTED: carbamoyl-phosphate synthase large chain, chloroplastic [Jatropha curcas] |
| 8 |
Hb_005854_040 |
0.0967084435 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_002392_020 |
0.1009407259 |
- |
- |
PREDICTED: UDP-N-acetylglucosamine--dolichyl-phosphate N-acetylglucosaminephosphotransferase [Jatropha curcas] |
| 10 |
Hb_106890_010 |
0.1040791622 |
- |
- |
PREDICTED: probable starch synthase 4, chloroplastic/amyloplastic isoform X2 [Jatropha curcas] |
| 11 |
Hb_002828_060 |
0.1046641059 |
- |
- |
PREDICTED: probable phytol kinase 3, chloroplastic [Jatropha curcas] |
| 12 |
Hb_005914_040 |
0.1066244869 |
- |
- |
o-linked n-acetylglucosamine transferase, ogt, putative [Ricinus communis] |
| 13 |
Hb_001998_220 |
0.1090240464 |
- |
- |
PREDICTED: isoamylase 1, chloroplastic [Jatropha curcas] |
| 14 |
Hb_001226_130 |
0.1094848833 |
- |
- |
PREDICTED: aminopeptidase M1-like [Jatropha curcas] |
| 15 |
Hb_021297_020 |
0.10960289 |
- |
- |
Cyclic nucleotide-gated ion channel, putative [Ricinus communis] |
| 16 |
Hb_023988_030 |
0.1097518616 |
- |
- |
PREDICTED: transmembrane protein 19 [Vitis vinifera] |
| 17 |
Hb_000027_200 |
0.1099706319 |
- |
- |
PREDICTED: uncharacterized protein LOC105642055 [Jatropha curcas] |
| 18 |
Hb_000984_140 |
0.1110933863 |
- |
- |
PREDICTED: xylulose kinase [Jatropha curcas] |
| 19 |
Hb_001366_270 |
0.1111920253 |
- |
- |
PREDICTED: LRR receptor-like serine/threonine-protein kinase RPK2 [Jatropha curcas] |
| 20 |
Hb_000976_110 |
0.1112868688 |
- |
- |
PREDICTED: KRR1 small subunit processome component homolog [Jatropha curcas] |