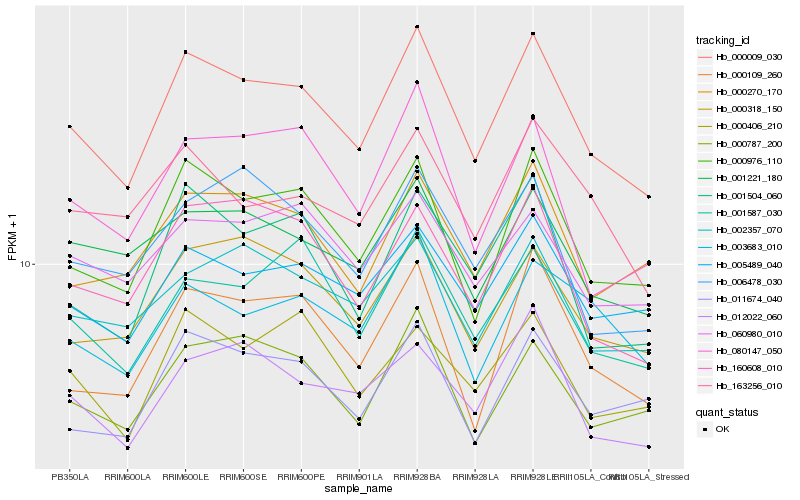
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000009_030 |
0.0 |
- |
- |
PREDICTED: splicing factor U2af large subunit B isoform X1 [Jatropha curcas] |
| 2 |
Hb_000976_110 |
0.0601895436 |
- |
- |
PREDICTED: KRR1 small subunit processome component homolog [Jatropha curcas] |
| 3 |
Hb_005489_040 |
0.0721487533 |
- |
- |
PREDICTED: cullin-1 [Jatropha curcas] |
| 4 |
Hb_000318_150 |
0.0753343371 |
- |
- |
RNA-binding protein Nova-1, putative [Ricinus communis] |
| 5 |
Hb_000787_200 |
0.0773657453 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 6 |
Hb_160608_010 |
0.0774065303 |
- |
- |
PREDICTED: BRASSINOSTEROID INSENSITIVE 1-associated receptor kinase 1-like isoform X1 [Populus euphratica] |
| 7 |
Hb_001587_030 |
0.0784981968 |
- |
- |
hypothetical protein JCGZ_09892 [Jatropha curcas] |
| 8 |
Hb_001221_180 |
0.079458741 |
- |
- |
calcineurin-like phosphoesterase [Manihot esculenta] |
| 9 |
Hb_000270_170 |
0.079666485 |
- |
- |
PREDICTED: formin-like protein 5 [Jatropha curcas] |
| 10 |
Hb_002357_070 |
0.0800713508 |
transcription factor |
TF Family: PHD |
PREDICTED: protein strawberry notch homolog 1 [Jatropha curcas] |
| 11 |
Hb_011674_040 |
0.0800976963 |
- |
- |
PREDICTED: uncharacterized protein LOC105648163 isoform X1 [Jatropha curcas] |
| 12 |
Hb_001504_060 |
0.0804970673 |
- |
- |
PREDICTED: nuclear pore complex protein NUP93A-like [Jatropha curcas] |
| 13 |
Hb_080147_050 |
0.0827069139 |
- |
- |
PREDICTED: probable plastidic glucose transporter 2 [Jatropha curcas] |
| 14 |
Hb_012022_060 |
0.0830295266 |
- |
- |
PREDICTED: uncharacterized protein LOC105646588 isoform X2 [Jatropha curcas] |
| 15 |
Hb_060980_010 |
0.0848691028 |
- |
- |
PREDICTED: BRASSINOSTEROID INSENSITIVE 1-associated receptor kinase 1-like isoform X1 [Populus euphratica] |
| 16 |
Hb_000109_260 |
0.0851615601 |
- |
- |
PREDICTED: WAT1-related protein At3g28050-like isoform X1 [Jatropha curcas] |
| 17 |
Hb_163256_010 |
0.0866782834 |
- |
- |
PREDICTED: phosphoglucomutase, chloroplastic [Jatropha curcas] |
| 18 |
Hb_000406_210 |
0.0868320852 |
- |
- |
PREDICTED: protein HASTY 1 [Jatropha curcas] |
| 19 |
Hb_003683_010 |
0.0880945647 |
- |
- |
longevity assurance factor, putative [Ricinus communis] |
| 20 |
Hb_006478_030 |
0.0891112622 |
- |
- |
hypothetical protein POPTR_0005s23510g [Populus trichocarpa] |