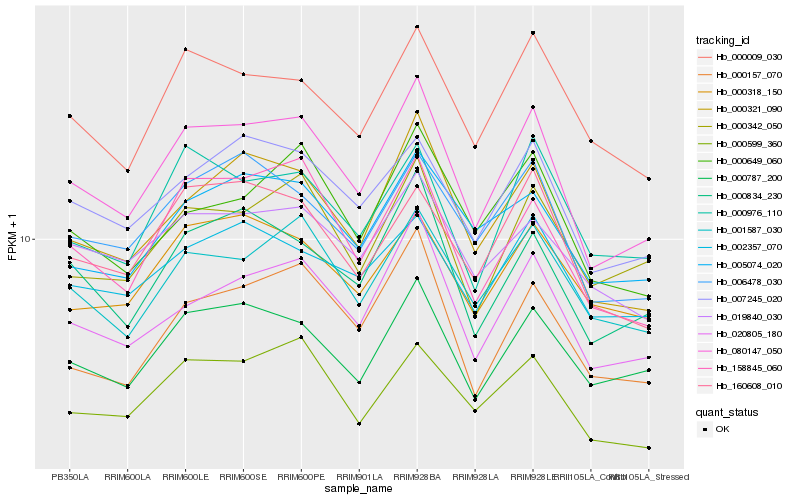
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_080147_050 |
0.0 |
- |
- |
PREDICTED: probable plastidic glucose transporter 2 [Jatropha curcas] |
| 2 |
Hb_020805_180 |
0.0651657818 |
- |
- |
PREDICTED: epidermal growth factor receptor substrate 15-like 1 [Jatropha curcas] |
| 3 |
Hb_000321_090 |
0.0658122308 |
- |
- |
PREDICTED: calcium-transporting ATPase 1, endoplasmic reticulum-type-like [Jatropha curcas] |
| 4 |
Hb_000834_230 |
0.0717538248 |
- |
- |
PREDICTED: ras-related protein RHN1-like [Jatropha curcas] |
| 5 |
Hb_000787_200 |
0.0730975036 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 6 |
Hb_158845_060 |
0.0739148172 |
- |
- |
PREDICTED: nudix hydrolase 3 isoform X1 [Jatropha curcas] |
| 7 |
Hb_007245_020 |
0.0741584811 |
- |
- |
PREDICTED: pre-mRNA-processing factor 17-like isoform X1 [Populus euphratica] |
| 8 |
Hb_001587_030 |
0.0776403485 |
- |
- |
hypothetical protein JCGZ_09892 [Jatropha curcas] |
| 9 |
Hb_006478_030 |
0.0778366845 |
- |
- |
hypothetical protein POPTR_0005s23510g [Populus trichocarpa] |
| 10 |
Hb_000342_050 |
0.0810501028 |
- |
- |
hypothetical protein POPTR_0006s24710g [Populus trichocarpa] |
| 11 |
Hb_000599_360 |
0.0812738254 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: putative disease resistance RPP13-like protein 1 [Jatropha curcas] |
| 12 |
Hb_160608_010 |
0.0823935535 |
- |
- |
PREDICTED: BRASSINOSTEROID INSENSITIVE 1-associated receptor kinase 1-like isoform X1 [Populus euphratica] |
| 13 |
Hb_000009_030 |
0.0827069139 |
- |
- |
PREDICTED: splicing factor U2af large subunit B isoform X1 [Jatropha curcas] |
| 14 |
Hb_000157_070 |
0.0853091348 |
- |
- |
PREDICTED: alpha-N-acetylglucosaminidase [Jatropha curcas] |
| 15 |
Hb_000976_110 |
0.0862761331 |
- |
- |
PREDICTED: KRR1 small subunit processome component homolog [Jatropha curcas] |
| 16 |
Hb_000649_060 |
0.086773073 |
- |
- |
PREDICTED: E3 ubiquitin protein ligase RIE1 [Jatropha curcas] |
| 17 |
Hb_019840_030 |
0.0886295285 |
- |
- |
PREDICTED: KH domain-containing protein At4g18375 isoform X2 [Jatropha curcas] |
| 18 |
Hb_005074_020 |
0.089844253 |
- |
- |
PREDICTED: uncharacterized protein LOC105644455 isoform X1 [Jatropha curcas] |
| 19 |
Hb_002357_070 |
0.0902718924 |
transcription factor |
TF Family: PHD |
PREDICTED: protein strawberry notch homolog 1 [Jatropha curcas] |
| 20 |
Hb_000318_150 |
0.090425998 |
- |
- |
RNA-binding protein Nova-1, putative [Ricinus communis] |