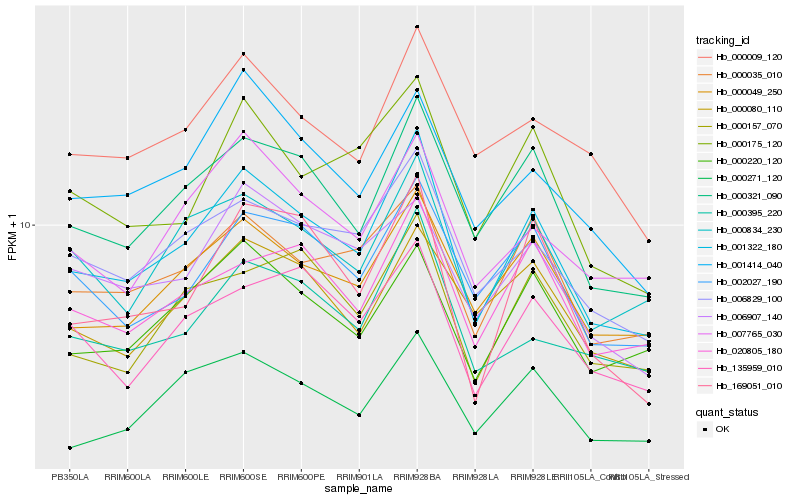
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001322_180 |
0.0 |
transcription factor |
TF Family: C2C2-GATA |
PREDICTED: GATA transcription factor 26 [Jatropha curcas] |
| 2 |
Hb_000321_090 |
0.0731377498 |
- |
- |
PREDICTED: calcium-transporting ATPase 1, endoplasmic reticulum-type-like [Jatropha curcas] |
| 3 |
Hb_000049_250 |
0.084825513 |
- |
- |
PREDICTED: uncharacterized protein LOC105644455 isoform X1 [Jatropha curcas] |
| 4 |
Hb_006829_100 |
0.0849168281 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_000834_230 |
0.0885258727 |
- |
- |
PREDICTED: ras-related protein RHN1-like [Jatropha curcas] |
| 6 |
Hb_000035_010 |
0.090647695 |
transcription factor |
TF Family: ARR-B |
PREDICTED: two-component response regulator ARR12-like isoform X1 [Jatropha curcas] |
| 7 |
Hb_002027_190 |
0.0918488111 |
- |
- |
PREDICTED: autophagy-related protein 18g [Jatropha curcas] |
| 8 |
Hb_000395_220 |
0.0951310869 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: ultraviolet-B receptor UVR8 [Jatropha curcas] |
| 9 |
Hb_000175_120 |
0.095180641 |
- |
- |
PREDICTED: pre-mRNA-processing-splicing factor 8 [Jatropha curcas] |
| 10 |
Hb_020805_180 |
0.0971473014 |
- |
- |
PREDICTED: epidermal growth factor receptor substrate 15-like 1 [Jatropha curcas] |
| 11 |
Hb_000220_120 |
0.0988255549 |
- |
- |
PREDICTED: ATP-dependent RNA helicase DHX36 isoform X2 [Jatropha curcas] |
| 12 |
Hb_007765_030 |
0.0993468602 |
- |
- |
PREDICTED: neutral ceramidase [Jatropha curcas] |
| 13 |
Hb_001414_040 |
0.1025063078 |
- |
- |
PREDICTED: RNA-dependent RNA polymerase 1-like isoform X1 [Jatropha curcas] |
| 14 |
Hb_000009_120 |
0.1025265953 |
- |
- |
PREDICTED: probable Xaa-Pro aminopeptidase P isoform X1 [Jatropha curcas] |
| 15 |
Hb_000157_070 |
0.1026884495 |
- |
- |
PREDICTED: alpha-N-acetylglucosaminidase [Jatropha curcas] |
| 16 |
Hb_169051_010 |
0.1038983203 |
- |
- |
JHL06B08.1 [Jatropha curcas] |
| 17 |
Hb_135959_010 |
0.1057378289 |
- |
- |
hypothetical protein JCGZ_07060 [Jatropha curcas] |
| 18 |
Hb_000080_110 |
0.1070840779 |
- |
- |
PREDICTED: GPI ethanolamine phosphate transferase 3 isoform X1 [Jatropha curcas] |
| 19 |
Hb_006907_140 |
0.1086808503 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_000271_120 |
0.1093207348 |
- |
- |
PREDICTED: U-box domain-containing protein 13-like [Nelumbo nucifera] |