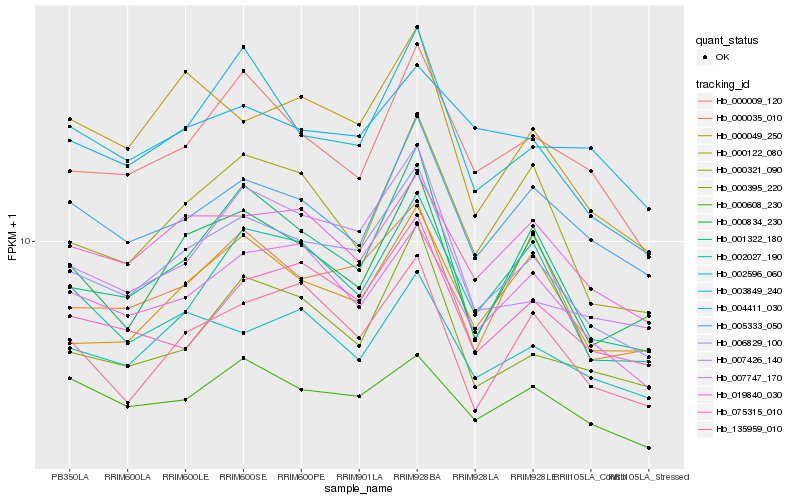
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_006829_100 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_007747_170 |
0.0848473672 |
- |
- |
PREDICTED: uncharacterized protein At5g41620 [Jatropha curcas] |
| 3 |
Hb_001322_180 |
0.0849168281 |
transcription factor |
TF Family: C2C2-GATA |
PREDICTED: GATA transcription factor 26 [Jatropha curcas] |
| 4 |
Hb_007426_140 |
0.0863955709 |
- |
- |
ornithine aminotransferase, putative [Ricinus communis] |
| 5 |
Hb_019840_030 |
0.0868222521 |
- |
- |
PREDICTED: KH domain-containing protein At4g18375 isoform X2 [Jatropha curcas] |
| 6 |
Hb_000009_120 |
0.0883018728 |
- |
- |
PREDICTED: probable Xaa-Pro aminopeptidase P isoform X1 [Jatropha curcas] |
| 7 |
Hb_000834_230 |
0.0886686372 |
- |
- |
PREDICTED: ras-related protein RHN1-like [Jatropha curcas] |
| 8 |
Hb_000035_010 |
0.0914696233 |
transcription factor |
TF Family: ARR-B |
PREDICTED: two-component response regulator ARR12-like isoform X1 [Jatropha curcas] |
| 9 |
Hb_000049_250 |
0.0916719456 |
- |
- |
PREDICTED: uncharacterized protein LOC105644455 isoform X1 [Jatropha curcas] |
| 10 |
Hb_002596_060 |
0.0917683282 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_005333_050 |
0.0919922487 |
- |
- |
PREDICTED: glucose-6-phosphate isomerase 1, chloroplastic [Jatropha curcas] |
| 12 |
Hb_075315_010 |
0.0927669671 |
- |
- |
ornithine aminotransferase [Camellia sinensis] |
| 13 |
Hb_000321_090 |
0.0938611258 |
- |
- |
PREDICTED: calcium-transporting ATPase 1, endoplasmic reticulum-type-like [Jatropha curcas] |
| 14 |
Hb_004411_030 |
0.0938761554 |
- |
- |
hypothetical protein POPTR_0017s00310g [Populus trichocarpa] |
| 15 |
Hb_000122_080 |
0.0943364813 |
- |
- |
PREDICTED: staphylococcal-like nuclease CAN2 [Jatropha curcas] |
| 16 |
Hb_000395_220 |
0.0961099312 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: ultraviolet-B receptor UVR8 [Jatropha curcas] |
| 17 |
Hb_002027_190 |
0.0971740399 |
- |
- |
PREDICTED: autophagy-related protein 18g [Jatropha curcas] |
| 18 |
Hb_003849_240 |
0.0974847672 |
- |
- |
leucine-rich repeat-containing protein, putative [Ricinus communis] |
| 19 |
Hb_000608_230 |
0.0977324669 |
- |
- |
PREDICTED: putative pentatricopeptide repeat-containing protein At1g12700, mitochondrial [Jatropha curcas] |
| 20 |
Hb_135959_010 |
0.098281648 |
- |
- |
hypothetical protein JCGZ_07060 [Jatropha curcas] |